ABSTRACT
DNA vaccination holds the potential to treat or prevent nearly any immunogenic disease, including cancer. To date, these vaccines have demonstrated limited immunogenicity in vivo due to the absence of a suitable delivery system which can protect DNA from degradation and improve transfection efficiencies in vivo. Recently, microneedles have been described as a novel physical delivery technology to enhance DNA vaccine immunogenicity. Of these devices, dissolvable microneedles promise a safe, pain-free delivery system which may simultaneously improve DNA stability within a solid matrix and increase DNA delivery compared to solid arrays. However, to date little work has directly compared the suitability of different dissolvable matrices for formulation of DNA-loaded microneedles. Therefore, the current study examined the ability of 4 polymers to formulate mechanically robust, functional DNA loaded dissolvable microneedles. Additionally, complexation of DNA to a cationic delivery peptide, RALA, prior to incorporation into the dissolvable matrix was explored as a means to improve transfection efficacies following release from the polymer matrix. Our data demonstrates that DNA is degraded following incorporation into PVP, but not PVA matrices. The complexation of DNA to RALA prior to incorporation into polymers resulted in higher recovery from dissolvable matrices, and increased transfection efficiencies in vitro. Additionally, RALA/DNA nanoparticles released from dissolvable PVA matrices demonstrated up to 10-fold higher transfection efficiencies than the corresponding complexes released from PVP matrices, indicating that PVA is a superior polymer for this microneedle application.
Introduction
DNA vaccines have been demonstrated to induce highly specific and potent humoural and cellular immune responses against pathogens and antigen-expressing tumors.Citation1,2 In addition, increased stability, ease of production and no risk of reversion to virulence have made DNA vaccines an attractive alternative to their conventional counterparts.Citation1 Despite their potential, DNA vaccines have suffered from limited immunogenicity, owing to a lack of suitable delivery system capable of overcoming the barriers to gene delivery. DNA vaccines rely on the delivery of antigen-coding DNA to the nucleus of cells, but due to the large molecular weight, anionic charge and susceptibility of DNA to enzymatic degradation physical injection of DNA often results in very low transfection efficacies.Citation3 Physical methods of disrupting the cell membrane such as electroporationCitation4 and ballistic deliveryCitation5 have been demonstrated to enhance gene expression levels significantly in vivo. However these devices are associated with pain,Citation6,7 and require the use of expensive specialist equipment, making them less acceptable to patients and unsuitable for mass vaccination. Alternatively, complexation of DNA with cationic lipids, polymers and peptidesCitation8,9 improves transfection efficacies in vivo via condensation of DNA into the nanoparticle range, improving cell uptake. While effective, these vectors have been limited by significant toxicity problems.Citation10
One strategy to enhance DNA vaccine potency is to deliver DNA into the highly immunogenic layers of the skin which harbours a wealth of antigen-presenting cells (APCs).Citation11 One of the predominant functions of the skin is the exclusion and detection of pathogens,Citation12 and the skin harbours a wealth of epidermal (known as Langherans cells [LCs]) and dermal dendritic cells (DCs) which serve as a link between the innate and adaptive branches of immunity.Citation13-15 Compared to other “professional” APCs such as macrophages and B cells, DCs are especially equipped for T cell activation, owing to the high quantity of co-stimulatory ligands at the DC surface which bind to co-stimulatory receptors proximal to the T Cell Receptor and thus are prime targets for DNA vaccination.Citation16,17 To this end microneedles are being investigated as a gene delivery tool.Citation18 Microneedles can be defined as a series of sharp microprojections, ranging up to 1000 µm in length, which are capable of piercing the outer barrier of the skin, the stratum corneum (SC), to create transient pores to the viable epidermal and dermal layers.Citation19 Microneedle-mediated DNA vaccines have demonstrated efficacy in mouse models against pathogenic diseases, such as hepatitis BCitation20 and influenza,Citation21 and perhaps more impressively against antigen-expressing tumors.Citation22,23 Initial gene delivery studies focused on applying DNA solutions to the skin, followed by application of solid microneedles,Citation24,25 however variability in dosing and high wastage of cargo have seen a shift toward the use of coated microneedles. Coating solid silicon or metal microneedles with rapidly dissolving DNA solutions enables a more consistent cargo loading and increased stability, however cargo delivery is limited to small quantities of DNA in the coating. Repeated drip-drying cycles may be used to increase the quantity of DNA present in coatings, however this is cumbersome and may affect needle conformation, and hence the ability to penetrate the SC.Citation21,26 Dissolvable microneedles have the potential to overcome these issues and deliver larger quantities of DNA, as cargo may be distributed throughout the whole needle as opposed to the coating.Citation27 Once applied, these structures swell within the interstitial fluid in the skin and undergo rapid dissolution to release their payload, thus it is essential that the needles are composed of biocompatible material and possess sufficient strength in the dry state to penetrate the SC. Importantly, novel fabrication processes for polymeric needles have been described to improve the scalability, and hence accessibility, of these devices.Citation27,28 For example, Lutton et al (2015), reported on a novel scalable manufacturing process for hydrogel needles.Citation28 Using a combination of injection molding and roller casting, the authors were able to fabricate highly reproducible polymeric microneedles at ambient temperatures and low cost. Arrays possessed similar characteristics to microneedles produced using their previously reported centrifugation method, but avoided the labor intensive process.Citation28 Hence, the limitations of small scale batch manufacturing methods currently used for microneedle fabrication within the laboratory, such as the time-consuming nature and material wastage,Citation29 are likely to be overcome when moving to production at an industrial scale using similar systems.
Although dissolvable microneedles have been examined extensively as drug delivery tools there have only been a few reports of dissolvable microneedles being used for nucleic acid delivery in vivo.Citation20,29-31 The stability of inactivated viruses and protein antigens has been established in certain dissolvable microneedle formulationsCitation32-34 however, little investigation has been done to determine whether incorporation into dissolvable microneedles has any influence on the stability of DNA or to directly compare the suitability of different polymer matrices to deliver nucleic acid cargo. This is perhaps surprising given that some polymers have been known to form hydrogen bonds with DNA and have been reported to be capable of enhancing gene delivery in their own right.Citation35 Therefore the current study is designed to determine the optimal polymer for delivery of nucleic acid cargo. Four different polymers were used to form dissolvable microneedles, loaded with or without nucleic acid cargo. The effect of needle composition on mechanical properties, the ability to release DNA and the functional stability of the DNA following release from the polymer matrices is reported. In addition, the ability of a novel amphipathic peptide, RALA,Citation36 to improve DNA recovery and transfection efficacies following release from polymer matrices is investigated.
Materials and methods
Materials
RALA peptide
The RALA peptide was purchased from Biomatik (USA) and supplied as an acetate salt in lyophilised form. The RALA peptide was stored at −20°C and reconstituted in DNase/RNase Free Water (Life Technologies, UK) immediately prior to use.
Plasmids
Reporter plasmids coding green fluorescent protein (GFP), pEGFP-N1, and luciferase, pCMV-Red Fire-fly Luc, were purchased from Clontech (USA) and Addgene (USA) respectively. Plasmids were transformed into and amplified in competent Escherichia coli DH5α cells (Life Technologies, UK). Prior to use plasmids were isolated and purified from transformed DH5α cells using PureLink HiPure Plasmid Filter Maxiprep Kits (Life Technologies, UK). DNA purity and concentration were determined according to UV absorbance at 260 and 280 nm (NanoDrop 2000c, Thermo Scientific, UK).
Dissolvable polymers
Polyvinylpyrrolidone (PVP) with molecular weight 58,000 Da (PVP-58) and 360,000 Da (PVP-360) were purchased from Ashland Inc. (USA) and Sigma Aldrich (UK) respectively. Polyvinyl alcohol (PVA) with molecular weight 13–23,000 Da (PVA-13-23) and 9-10,000 Da (PVA-9-10) were both purchased from Sigma Aldrich (UK).
Cell line and cell culture
NCTC 929 murine fibroblast cells were purchased from American Type Culture Collection (USA) and maintained in Minimum Essential Media (MEM) (Life Technologies, UK) supplemented with 10% Foetal Horse Serum (FHS) (Life Technologies, UK) at 37°C with 5% CO2. Opti-MEM (Life Technologies, UK) was used for cell transfection protocols.
Other reagents
MTS cell proliferation assay media, CellTiter 96 Aqueous One Solution Reagent, was purchased from Promega (USA). D-Luciferin was purchased from Gold Biotechnology (USA), Proteinase K and Tris(hydroxymethyl)aminomethane were purchased from Sigma Aldrich (UK). Primers for qRT-PCR were purchased from Integrated DNA Technologies (UK). Quanti-iT Picogreen Reagent was purchased from Life Technologies (UK).
Animals
Male C57BL/6 mice, 6–8 weeks old, were purchased from Harlan UK Ltd (UK). Mice were housed in an open facility with unlimited access to food and water. All experimental procedures were carried out under project license 2794 and complied with the UK Scientific Act 1986.
Preparation of RALA/DNA nanoparticles
RALA/pDNA nanoparticles were prepared at varying concentrations at the desired N:P ratio (the ratio of the positively charged nitrogen atoms from the peptide to the negatively charged phosphates on the DNA backbone) as described elsewhere.Citation36 Briefly, the RALA peptide solution was added to DNA solutions and nanoparticles were left to incubate for 30 min at room temperature prior to use.
Manufacture of dissolvable microneedle arrays
Silicon microneedle molds, prepared as described previously,Citation37 were used as a template for microneedle formulation. Arrays consisted of 361 (19×19) conically shaped needles measuring 600 µm in length, with a base width of 300 µm and 50 µm spacing.
To prepare cargo-free arrays approximately 500 mg of 20% w/w (PVP-360, PVA-13-23 and PVA-9-10) or 300 mg of 30% w/w (PVP-58) aqueous polymer solution was added to micromoulds, centrifuged for 10 min at 4,000 rpm and allowed to dry at room temperature for 48 h prior to use. Following 24 h of drying at room temperature 500 mg of PVP-360 was added to arrays fabricated from PVP-58 and centrifuged for 10 min at 4,000 rpm.
To prepare RALA/pDNA loaded arrays, RALA/pDNA solutions were mixed at a ratio of 60:40 with concentrated aqueous polymer stock solutions (50% w/w for PVP-360, PVA-13-23 and PVA-9-10 and 75% w/w for PVP-58). To prevent wastage of DNA cargo, 25 mg of aqueous polymer-RALA/pDNA solutions were then added to the micromoulds and centrifuged for 10 mins at 4,000 rpm to fill the micromould indents. To form an inert supporting baseplate 500 mg of 20% w/w (PVP-360, PVA-13-23 and PVA-9-10) or 200 mg of 30% w/w (PVP-58) aqueous polymer solution was added to the moulds and centrifuged for 10 min at 4,000 rpm. Following 24 h of drying at room temperature 500 mg of PVP-360 was added to arrays fabricated from PVP-58 and centrifuged for 10 min at 4,000 rpm. Arrays were then left to dry at room temperature for a further 24 h prior to use.
Following drying, arrays were carefully peeled from moulds and sidewalls were removed with a heated scalpel immediately prior to use.
Visual assessment of needle characteristics
Prior to imaging, microneedle arrays were adhered to a flat steel block using double-sided tape and positioned perpendicular to the microscope. Arrays were imaged using at x35 magnification using a Leica EZ4D digital microscope (Leica, Germany) and needle lengths were measured using inbuilt microscope software.
Mechanical testing of microneedle arrays
Mechanical properties of microneedles were assessed using a TA-XT2 Texture Analyser (Stable Microsystems, UK). Microneedle arrays were measured, adhered with double-sided tape to the movable probe of the TA-XT2 Texture Analyser and subjected to a compression force for 45 N (0.125 N/needle) for 30 s. Following compression, needles were re-measured and percentage height reduction was calculated as the difference in microneedle height following compression divided by the original height multiplied by 100.
Microneedle array penetration into neonatal porcine skin
Microneedle penetration into the skin was assessed using optical coherence tomography (OCT). Neonatal porcine skin was used as a model for penetration studies. Skin was stored at −20°C between experiments. Immediately prior to analysis skin was thawed in PBS at 37°C for 30 min and carefully shaved using a disposable razor. Skin was placed, SC side facing up, on to a sheet of dental wax for support and DNA free-microneedles were then inserted into the skin using manual pressure, and held in place for 30 s. Following insertion, images of needle penetration were obtained using an EX1301 OCT microscope (Michelson Diagnostics Ltd, UK). Image J software (National Institutes of Health, USA) was then used to analyze 2D images of microneedle penetration into skin. Accurate measurements of needle penetration depths were obtained using the scale of the images where 1.0 pixel = 4.2 µm, and percentage penetration depth was assessed as needle length inserted into the skin divided by the total needle length multiplied by 100. The microneedle penetration volumes were subsequently determined using the equation V = π x r2 x (h/3), where r is the microneedle radius at the point of penetration, and h is the length of microneedle inserted into the skin. The percentage penetration volume was determined as the volume of needle inserted in the skin divided by the total needle volume multiplied by 100.
Assessment of cell viability following exposure to polymers matrices
The affect of exposure to polymer matrices was assessed using the MTS cell proliferation assay in the NCTC 929 fibroblast cell line. This cell line has been previously used to assess the biocompatibility of dissolvable microneedle formulations,Citation29 and is listed by ATCC as being suitable for toxicity testing and transfection studies (www.lgcstandards-atcc.org/products/all/CCL-1). NCTC 929 fibroblast cells were seeded into 96 well plates at densities of 10,000 cells/well and left to adhere overnight. The following day polymers were added to wells at concentrations of 0–40 mg/mL in complete media and cells were incubated for a further 24 h. After this time, media was removed and replaced with 200 µL/well of fresh complete media and supplemented with 10% CellTiter 96 Aqueous One Solution Reagent. Cells were incubated for a further 2 h at 37°C and 5% CO2 and the absorbance at 490 nm was subsequently measured using an EL808 96-well plate reader (BioTek Instruments Inc., UK). Control cells, not exposed to any polymer, were taken to be 100% viable and the cell viability of cells exposed to increasing concentrations of polymer was determined as a percentage of the absorbance of untreated control cells.
DNA recovery and integrity analysis following incorporation into the dissolvable polymer matrices
To avoid wastage of DNA and the time-consuming process of manufacturing microneedles, the effect of polymer matrices on DNA recovery, integrity and transfection efficacies was determined using polymer gels manufactured in silicon micromoulds lacking needle indents.
For DNA recovery experiments 250 mg polymer gels containing 10 µg of pEGFP-N1 were prepared by mixing RALA/pEGFP-N1 solutions (N:P 0–10) at a ratio of 60:40 with concentrated aqueous polymer stock solutions (50% w/w for PVP-360, PVA-13-23 and PVA-9-10 and 75% w/w for PVP-58). Following drying polymer gels were completely dissolved in TRIS buffer (pH 8) and samples were incubated with Proteinase K 1.0 mg/mL solution for 2 h at 37°C to allow dissociation of pDNA from the RALA peptide prior to analysis.Citation38 pDNA release was quantified using the Quanti-iT Picogreen Assay.
For DNA integrity experiments polymer gels containing 30 µg of pEGFP-N1 (N:P 0 and 6) were fabricated as above. Following drying, polymer gels were completely dissolved in TRIS buffer (pH 8) and samples were incubated in TRIS buffer (pH 8) with or without Proteinase K 1.0 mg/mL solution for 2 h at 37°C. Samples were then electrophoresed for 1 h on a 1% agarose gel incorporating Ethidium Bromide which was then imaged using the Multispectrum Bioimaging System (UVP, UK).
In vitro transfection analysis following release from the polymer matrices
For DNA transfection protocols polymer gels were prepared as above containing 20 µg of pEGFP-N1 (N:P 0-12) and left to dry at room temperature for 48 h. NCTC 929 fibroblast cells were seeded into 24 well plates at densities of 100,000 cells/well and left to adhere overnight. The following morning complete media was removed from wells and replaced with 500 µL Opti-MEM reduced serum media for 2 h. Dried polymer gels were dissolved in 1,000 µL of Opti-MEM media for 1 h and subsequently 250 µL/well samples were incubated with NCTC 929 cells for 6 h at 37°C and 5% CO2. Following this time the cell supernatant was removed and replaced with 500 µL of complete media. Cells were then incubated for a further 48 h at 37°C and 5% CO2 prior to analysis of transfection efficacy.
To visualize GFP expression following transfection, cells were imaged at x10 magnification using the EVOS FL Cell Imaging System (Thermofisher Scientific, UK). Following imaging, cells were trypsinised, harvested in 2% Formaldehyde and stored at 4°C prior to flow cytometric analysis. GFP expression was detected using the FACS Calibur System (BD Bioscience, UK) and the data was analyzed using Cyflogic software (CyFlo Ltd, Finland).
Quantification of DNA recovery and delivery from RALA/DNA loaded microneedles
Dissolvable microneedle arrays were formulated with 32 µg of pDNA by dilution of concentrated polymer stock with RALA/pDNA (N:P 10) aqueous solution, as described previously. The quantity of DNA residing in the microneedle array following manufacture was determined using the Picogreen assay as previously described. Sidewalls were removed prior to experimentation to allow separate analysis from arrays (comprised of baseplate and microneedles).
Following determination of RALA/pDNA loading within dissolvable microneedle patches, the quantity of pDNA delivered in vivo from arrays was determined. Dissolvable microneedles were formulated using 32 µg of pDNA complexed into RALA/pDNA (N:P 10) nanoparticles as above, and sidewalls were removed prior to experimentation. Female C57/BL6 mice were anaesthetised using 5% Isofluorane in an induction chamber. Following induction, anesthesia was maintained using 3% Isofluorane delivered via a face mask during microneedle application. Microneedle arrays were applied into the dorsal side of mouse ears using manual application, and held in place for 5 min with gentle pressure. Subsequently, arrays were held in place for a further 24 h using 3M surgical tape (Micropore Ltd, UK), prior to removal to allow quantification of pDNA remaining in the microneedle patch using the Picogreen assay. The quantity of pDNA delivered to mouse ears was calculated by subtracting the quantity of pDNA remaining in microneedle arrays following application to mouse ears from the quantity of pDNA in unapplied microneedle arrays.
Results
Microneedle formulation, characterization and skin insertion studies
Dissolvable microneedles were successfully formed by simple centrifugation of a range of aqueous polymer solutions (PVP-360, PVP-58, PVA-13-23 and PVA-9-10) into silicon micromoulds (). The baseplate of arrays formulated from PVP-58 cracked upon peeling from the mastermould, necessitating the addition of less brittle PVP-360 to form a secondary baseplate layer. To determine whether the polymer formulation significantly affected the mechanical strength of needles, arrays were subjected to a 45 N axial compression and reimaged to determine the percentage height reduction. No needle fracture was apparent following compression and in addition, needle conformation was largely unchanged (), although a slight blunting of the tip ends was observed. Needles composed of PVA-9-10 underwent the greatest reduction in height (10.237% ± 0.807), although this was not significantly higher than arrays composed of PVA-13-23, which resisted compression most effectively (7.077% ± 1.446, p > 0.05) ().
Figure 1. Dissolvable microneedle arrays produced by simple centrifugation are able to withstand high compression forces and penetrate the stratum corneum. (A) Light microscope images of microneedles formulated from i) 360 kDa PVP, ii) 58 kDa PVP, iii) 13–23 kDa PVA and iv) 9–10 kDa PVA prior to and (B) following 45 N axial compression; (C) Percentage height reduction of needles following 45 N compression (means + SEM, N = 3); (D) Optical Coherence Tomography (OCT) images of i) 360 kDa PVP, ii) 58 kDa PVP, iii) 13–23 kDa PVA and iv) 9–10 kDa PVA microneedle penetration into neonatal porcine skin following application of manual pressure and (E) the corresponding percentage penetration of arrays through the stratum corneum (means + SEM, N = 3).
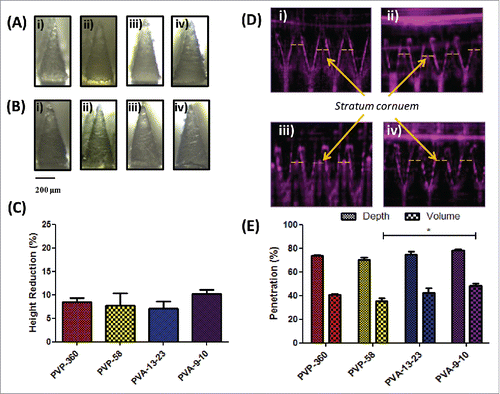
The functional ability of the dissolvable microneedles to breach the SC was then next assessed using OCT. Arrays were applied into the SC facing side of neonatal porcine skin using manual pressure and immediately imaged. Cross-sectional images of microneedles following application revealed a highly consistent, clear penetration through the SC () where needle penetration depths varied from 78.50% ± 0.956 (PVA-9-10) to 70.39% ± 1.837 (PVP-58) (p > 0.05), corresponding to between 48.53% ± 2.79 (PVA-9-10) to 35.13% ± 1.80 (PVP-58) (p < 0.05) of the total needle volume being inserted into the skin ().
Cytotoxicity assays
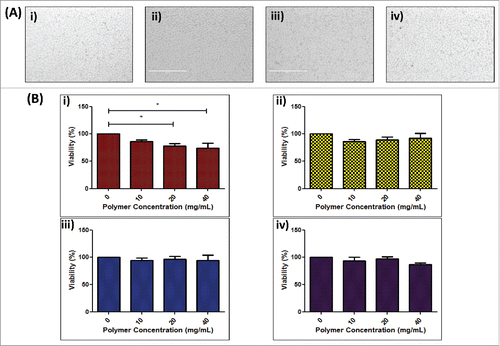
To investigate the potential toxicity of polymers to cells, NCTC-929 cells were exposed to dissolved DNA-loaded polymers and examined following 24 h for signs of stress or apoptosis (). Following exposure, cells maintained their conformation, showed no signs of shrinkage, membrane damage or blebbing, key morphological signs of apoptosis,Citation39 indicating that exposure to polymer matrices did not result in cell damage. The effect of increasing concentrations of polymers (PVP-360, PVP-58, PVA-13-23 and PVA-9-10) on cell viability was then examined via MTS assay (). The viability of cells was not significantly affected by exposure to PVP-58, PVA-13-23 or PVA-9-10 for 24 h (p < 0.05) and did not drop below 80%, even at concentrations up to 40 mg/mL. Conversely, cell viability decreased with exposure to increasing concentrations of PVP-360, with concentrations of 20 mg/mL and 40 mg/mL causing a significant decrease in cell viability to below 80% (p < 0.05).
Figure 2. Effect of polymer formulation on NCTC 929 cell viability. (A) Digital microscope of images of NCTC 929 fibroblast cells 24 h following exposure to dissolved DNA loaded i ) 360 kDa PVP, ii) 58 kDa PVP, iii) 13–23 kDa PVA and iv) 9–10 kDa PVA polymer matrices; (B) Cell viability of NCTC 929 cells following 24 h exposure to increasing concentrations of i ) 360 kDa PVP, ii) 58 kDa PVP, iii) 13–23 kDa PVA and iv) 9–10 kDa PVA polymer, expressed as percentage viability compared to untreated control (means + SEM, n = 3).
DNA recovery and stability within polymer formulations
To determine the quantity of pDNA that could be recovered from the respective polymer matrices polymer gels were formulated with known quantities of pDNA (10 µg) complexed to increasing amounts of RALA peptide. Polymer gels were then dissolved and the quantity of DNA released was determined via Picogreen assay. The quantity of pDNA recovered from PVA matrices was maximal across the range of N:P ratios examined (). Conversely, at N:P 0 (naked pDNA) the quantity of pDNA recovered from PVP matrices was 7.293 ± 1.240 µg and 7.741 ± 0.2119 µg from PVP-360 and PVP-58 respectively. pDNA recovery from PVP matrices increased with N:P ratio, up to N:P 4 when all of the loaded pDNA was recovered ().
Figure 3. The effect of polymer matrices on DNA recovery and integrity. (A) Recovery of DNA following incorporation into i) 360 kDa PVP, ii) 58 kDa PVP, iii) 13–23 kDa PVA and iv) 9–10 kDa PVA polymer gels at various N:P ratios (N:P 0–10) (mean + SEM, n = 3); (B) Gel electrophoresis of DNA (N:P 0) and RALA/DNA nanoparticles (N:P 6) following dissolution from polymer matrices following storage at room temperature for i) 0 or ii) 7 d. Lane 1: 1 kB DNA ladder, Lane 2: control DNA Lanes 3–6: DNA (N:P 0) recovered from 360 kDa PVP, 58 kDa PVP, 13–23 kDa PVA and 9–10 kDa PVA polymer gels incubated with Proteinase K, Lanes 7 – 10: RALA/DNA nanoparticles (N:P 6) recovered from 360 kDa PVP, 58 kDa PVP, 13–23 kDa PVA and 9–10 kDa PVA polymer gels incubated with Proteinase K, Lanes 11 – 14: RALA/DNA nanoparticles (N:P 6) recovered from 360 kDa PVP, 58 kDa PVP, 13–23 kDa PVA and 9–10 kDa PVA polymer gels incubated with TRIS buffer.
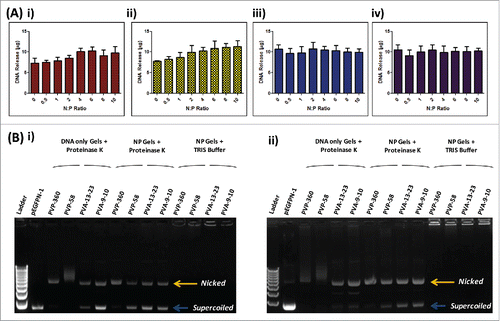
Gel electrophoresis was subsequently used to assess the stability of pDNA following incorporation into polymer formulations, either alone or complexed to RALA (N:P 6). Following incorporation into PVP formulations there was a clear loss of pDNA stability as the DNA conformation changes from predominantly supercoiled to nicked (). In addition, pDNA bands appear indistinct and smeared on the gel, supporting that at lower N:P ratios pDNA recovery from PVP formulations was compromised by pDNA degradation. Naked pDNA released from PVA formulations also shows a change in pDNA tertiary structure, although a high proportion of the supercoiled conformation was retained and there is no evidence of DNA degradation (). Following incorporation into polymer matrices RALA/pDNA nanoparticles remain intact, as shown by the inability of DNA to run through the agarose gel (). This is because nanoparticles have an overall positive charge and so do not migrate through the agarose gel upon application of a current. Therefore, in order to visualize the pDNA conformation, nanoparticles were incubated with Proteinase K to digest the RALA peptide prior to analysis (). Complexing pDNA to RALA prior to incorporation into PVP formulations led to some preservation of the supercoiled conformation and protected pDNA from degradation within the matrix, although the majority of the pDNA remained in the nicked conformation (). Conversely, complexing pDNA into nanoparticles prior to incorporation into PVA formulations did not cause a significant change to pDNA stability (). These results illustrate that the RALA peptide is necessary to prevent DNA degradation within PVP matrices and that the pDNA stability is compromised upon drying within polymer formulations.
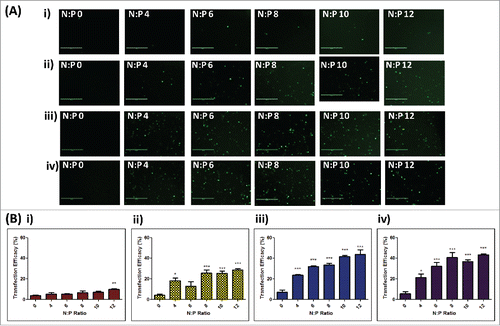
Figure 4. In vitro transfection efficacies of RALA/DNA nanoparticles at various N:P ratios (N:P 0–12) following dissolution from polymer matrices. (A) Representative fluorescent images and (B) cell transfection efficacies of NCTC 929 fibroblast cells following transfection with RALA/pEGFP-N1 nanoparticles released from i) 360 kDa PVP, ii) 58 kDa PVP, iii) 13–23 kDa PVA and iv) 9–10 kDa PVA polymer gels at various N:P ratios (N:P 0–12), (mean + SEM, n = 3).
Formulations were further incubated for 7 d at room temperature prior to investigating whether pDNA underwent further conformational changes following short-term storage. Further pDNA degradation is evident with naked pDNA released from PVP formulations (), and a further loss of the supercoiled pDNA conformation is evident from naked pDNA released from PVA formulations (). Complexing pDNA into nanoparticles led to a preservation of the pDNA structure with storage, as the proportions of pDNA in the supercoiled and nicked conformations remained largely unchanged after 7 d of storage ().
In vitro transfection efficacy
To assess whether pDNA remained functional following incorporation into polymer formulations, despite the change in structure, the transfection efficacy of pEGFP-N1 or RALA/pEGFP-N1 nanoparticles was determined in NCTC 929 cells following dissolution from loaded polymer gels. Cells incubated with naked pDNA demonstrated limited transfection following release from all polymers, however, as the quantity of RALA complexed to DNA (N:P ratio) increased, GFP expression due to transfection became more apparent (). Despite an increase in transfection efficacy, GFP expression following release of RALA/pEGFP-N1 nanoparticles from PVP-360 formulations remained limited throughout the range of N:P ratios (). Quantitative analysis of GFP transfection efficacies via flow cytometry was reflective of fluorescent images, demonstrating that significantly higher transfection efficacies were achieved with increasing N:P ratio compared to naked DNA (). The highest transfection efficacy achieved was 43.69% ± 4.598, following cell incubation with RALA/pEGFP-N1 nanoparticles (N:P 12) recovered from PVA-13-23 matrices. Similar transfection rates were seen following incubation with nanoparticles (N:P 12) released from PVA-9-10 formulations (43.35% ± 0.9778) (p > 0.05), however significantly lower transfection efficacies were achieved following release of nanoparticles (N:P 12) from PVP-360 (p = 0.0019) and PVP-58 (p = 0.0340) formulations. These results demonstrate that the functional integrity of pDNA has been compromised by incubation within PVP formulations. This loss of transfection efficacy may be related to the loss of DNA supercoiled conformation, which is greater within PVP formulations ().
Formulation and mechanical testing of DNA loaded microneedles
pDNA-loaded microneedles were formulated by centrifugation of aqueous polymer, diluted with pDNA or RALA/pDNA nanoparticle solution, to form needle tips prior to addition of an inert baseplate layer. To minimise the possibility of microneedle mechanical failure the concentration of polymer solution used for formulation remained consistent (20% PVP-360, PVA-13-23 and PVA-9-10, and 30% w/w for PVP-58). The quantity of pDNA that could be loaded into microneedle arrays was therefore limited by the workable concentrations of pDNA and RALA peptide which could be used for dilution of concentrated polymer stock. The concentrations of pDNA and RALA solutions used for microneedle formulation were 7.5 µg/mL and 43.5 µg/mL respectively. RALA/pDNA solutions at N:P 10 were chosen for mechanical strength comparisons which allowed a hypothetical pDNA loading of 32 µg per array ().
Figure 5. Characterization of dissolvable microneedle arrays loaded with DNA (N:(P)0) and RALA/DNA nanoparticles (N:(P)10). (A) Digital microscope images of microneedles formulated from i) 360 kDa PVP, ii) 58 kDa PVP, iii) 13–23 kDa PVA and iv) 9–10 kDa PVA loaded with RALA/DNA nanoparticles (N:P 10) prior to and (B) following 45 N axial compression; (C) Percentage height reduction of i) 360 kDa PVP, ii) 58 kDa PVP, iii) 13–23 kDa PVA and iv) 9–10 kDa PVA arrays, unloaded or loaded with DNA only(N:P 0) or RALA/DNA nanoparticles (N:P 10) following 45 N compression (means + SEM, N = 3).
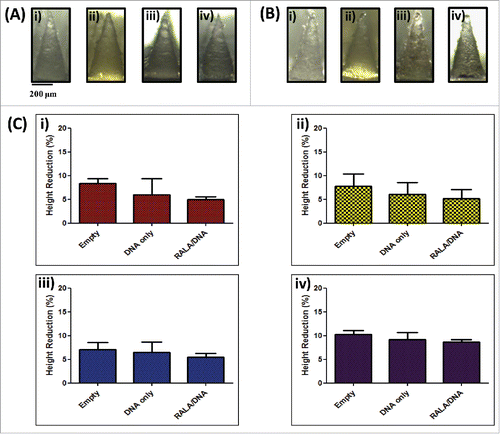
Arrays loaded with pDNA or RALA/pDNA nanoparticles were subjected to an axial compression force of 45 N and the needle percentage height reduction was calculated as previously described. Similarly to unloaded arrays, needles showed a blunting of tip ends () but underwent no major deformation or buckling. Incorporation of pDNA or nanoparticle cargo into arrays did not compromise needle mechanical properties, and none of the needles tested underwent a height reduction greater than 10% of the original needle length (). Therefore it was concluded that pDNA loaded needles would be able to disrupt the SC in a similar manner to unloaded microneedle arrays.
DNA recovery and delivery from loaded microneedle arrays
As it is likely that some pDNA shall be lost during the manufacturing process, the DNA loading in the baseplate and microneedles of arrays, was determined using the Picogreen assay. Microneedle arrays (baseplate and needles) composed of 20% w/w 9–10 kDa PVA were found to release the highest quantity of pDNA (17.7 µg), which was significantly greater than quantities of pDNA released from arrays composed of 360 kDa PVP (12.6 µg, p < 0.001), 58 kDa PVP (14.2 µg, p < 0.01) and 13–23 kDa PVA (15.4 µg, p < 0.05) as determined by one-way ANOVA ().
Figure 6. Determination of RALA/pDNA delivery from loaded dissolvable microneedle arrays in vivo. (A) Quantification of RALA/DNA encapsulation within the microneedle array baseplate and needles following drying and sidewall removal (B) Quantification of RALA/DNA encapsulation within removed microneedle sidewalls. (C) Quantification of RALA/DNA delivered from loaded microneedle arrays in vivo following application to mouse ears for 24 h.
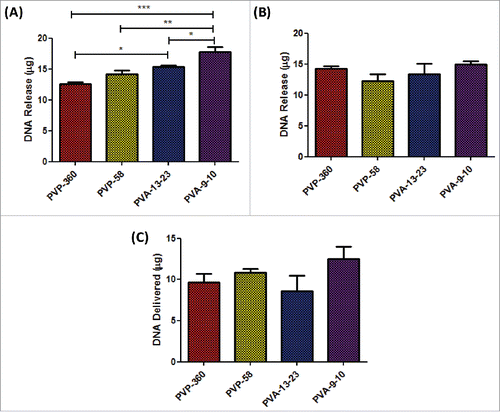
It was found that a large portion of pDNA was lost into the sidewalls for all microneedle formulations (), with 14.3 µg, 12.3 µg, 13.2 µg and 14.4 µg of pDNA being recovered from the sidewalls of 360 kDa PVP, 58 kDa PVP, 13–23 kDa PVA and 9–10 kDa PVA formulations respectively.
Following determination of the quantity of pDNA encapsulated into the baseplate and needles of microneedle arrays, the quantity of pDNA actually delivered from the patches in vivo was determined. Loaded microneedle arrays were applied to the dorsal side of mouse ears for 24 h and the pDNA remaining in the microneedle arrays following removal from mouse ears was determined. The quantity of pDNA delivered into mouse ears in vivo was subsequently estimated by subtracting the quantity of pDNA remaining from the pDNA loaded into the microneedle array prior to application (). The greatest quantity of pDNA delivered in vivo was 12.5 µg (17.7 μg – 5.2 µg) from the 9–10 kDa PVA formulations, although this was not significantly greater than the quantity of pDNA delivered from 360 kDa PVP (9.6 µg), 58 kDa PVP (10.9 µg) or 13–23 kDa PVA (8.6 µg) microneedle arrays.
Discussion
Recently, we reported on the development of a novel dissolvable 360 kDa PVP microneedle system incorporating cationic RALA/pDNA nanoparticles to aid transfection.Citation29 Although the recovered pDNA cargo was capable of eliciting gene expression in vitro and in vivo, transfection efficiency was reduced compared to fresh nanoparticles,Citation36 indicating a loss of functionality. Given that PVP has previously been reported to form hydrogen bonds with DNA, this loss in transfection efficacy was not entirely unexpected.Citation26 This then prompted us in this study to examine the suitability of 4 FDA-approved polymer formulations to form dissolvable microneedles for DNA vaccination. In addition, we incorporated the amphipathic cationic peptide RALA prior to formulation to enhance the functionality of the DNA cargo.
Firstly, it was established that all chosen formulations (PVP-360; PVP-58; PVA-13-23 and PVA-9-10) were equally suitable for microneedle fabrication. As with our previous study, we fabricated microneedles with 600 μm length in order to target DNA delivery to the epidermal and dermal layers of the skin. Previous work has demonstrated that utilizing this needle length results in microneedle penetration depths of 400–500 μm in pig skin, with no significant benefit observed with increasing needle length to 900 μm (penetration depths of 500–600 μm in the same model).Citation40 As the human epidermis differs in thickness from ∼70–180 μm this needle length is expected to fully penetrate through this layer into the upper layers of the dermis.Citation19,41 Therefore dissolution of the microneedle structure shall allow cargo to be targeted to both epidermal LCs and dermal DCs across a range of application sites.Citation13-15 Previous studies have reported that a lesser strength is one of the limitations of dissolvable microneedle matrices compared solid microneedles which can adversely affect penetration of the SC. Larrañeta et al (2014) determined that volunteers use a maximum average insertion force of 32 N when applying dissolvable microneedle patches.Citation40 In the above experiment 2 volunteers (of 20) were found to exert an average exertion force of ∼40 N when applying microneedles,Citation40 and therefore 45 N was selected as the application force that arrays must be capable of withstanding without significant deformation. Following a 45 N compression force, there was no significant difference in needle height reduction indicating that the formulations should have sufficient strength to withstand insertion (). Penetration of the SC is another critical factor to consider with dissolving microneedles and this is related to needle sharpness and strength of the formulating polymer.Citation19,27,42 Therefore the ability of microneedles to penetrate the SC was subsequently tested using neonatal porcine skin as a model for human skin. Penetration depths achieved in this study were approximately 70–80% of the needle length across the range of polymers tested (), which is similar to or greater than the penetration depths achieved using solid or coated needles.Citation26,30,43 Therefore we could be confident that a large proportion of the payload within the dissolvable polymer matrix would be deposited into the skin following microneedle insertion. The microneedle type can affect the outcomes of incomplete insertion. For example, coated microneedles will only deliver the cargo present on the inserted part of the needle.Citation26 In contrast, incomplete insertion of dissolvable needles may not be as limiting as dissolution of needles creates transient pores in the SC facilitating the passage of cargo from the un-inserted needle portion and the baseplate. This can be further enhanced when the interstitial fluid is absorbed and spreads through the polymer matrix during the dissolution process.Citation44 Given that all formulations in this study produced strong, sharp microneedles all 4 polymers displayed suitable characteristics for further in vitro analysis.
Given that the dissolvable matrix as well the cargo is deposited into the skin, the choice of polymer is also a critical factor. PVP has been shown to be almost completely eliminated through the kidneys at lower molecular weights < 25 kDa, which is approximately the limit of glomerular filtration in rat kidneys.Citation45 However, as the molecular weight of PVP increases, elimination is reduced and the PVP is retained in the reticulo-endothelial system,Citation46,47 and injection of PVP with molecular weight > 100 kDa has been shown to lead to storage disease.Citation48 As such, the high molecular weight of PVP-360, used by McCaffrey et al (2016) for microneedle fabrication,Citation29 may render it unsuitable for patient administration. Similarly, PVA elimination rates depend largely on the injection site and the molecular weight of the polymer, with the cut off size for glomerular filtration being 30 kDa.Citation49 Studies examining the fate of larger molecular weights of PVA have shown accumulation within the liver and spleen of rats.Citation50 From these collective studies it is clear that the molecular weight of the polymer should be as low as possible to ensure complete elimination. To date most studies have focused on intravenous administration of polymers and therefore in order to license a dissolvable microneedle product further investigation into the distribution and elimination following repeated intradermal (i.d.) administration will be crucial. Should the formulating polymer be retained in the skin for any period it is critical that no loss of skin integrity should occur. Currently no guidance has been provided from regulators with regards to the safety and toxicity standards which microneedles shall need to meet for licensure; however, the MTT and MTS viability assays, which measure cells' mitochondrial metabolic rate, have been used previously to determine the biocompatibility of dissolvable microneedle formulations.Citation29,51 Therefore we also examined the effect of increasing concentrations of polymer on cell viability in NCTC 929 fibroblast cells using the MTS viability assay. Results demonstrated that at concentrations up to 10 mg/mL, none of the polymers caused a significant decrease in cell viability. McCrudden et al (2014) found that exposure of L-132 cells to 5 mg/mL of MN formulation (PMVE/MA) resulted in greater than a 90% drop in cell viability yet this did not translate to significant loss of skin viability in a 3D model or irritation to rat skin in vivo following 24 h exposure to the MN formulation.Citation51 As such although higher concentrations of PVP-360 (20 mg/mL and 40 mg/mL) did result in a drop in cell viability below 80%, these concentration ranges are much higher than would be used in vivo, and are unlikely to result in damage to healthy skin.
At this point all 4 polymers were considered suitable to proceed with, however further studies revealed that pDNA release from the PVP polymer matrices was compromised. In this case we were able to achieve complete recovery of pDNA from PVA matrices, and from PVP matrices where pDNA was complexed with RALA peptide at N:P ratios >4 (). Quantification of pDNA release from polymer matrices is essential but gives little indication as to the integrity of the pDNA and further analysis by agarose gel revealed that pDNA was degraded within the PVP matrices. This loss of DNA integrity was not observed by Qiu et al (2015) within their dissolvable microneedle system, where pDNA functionality was determined via protein quantification following transfection with recovered pDNA.Citation52 One key difference may be that the solutions used for microneedle formulation in our study were a much higher concentration than those used by Qiu et al (2015) (5% w/w 10 kDa PVP) and thus the effect on pDNA stability may be more apparent.Citation52 Degradation of naked pDNA was greater in the more highly concentrated 58 kDa PVP matrix (30% w/w) than the 360 kDa PVP matrix (20% w/w) (). PVP has been previously reported to cause cleavage of DNA by hydrolysis of guanine and adenine bases.Citation53 During the formulation process it was observed that pDNA incorporated into PVA and PVP matrices also underwent a change in structure. To achieve high transfection efficacies retaining the supercoiled conformation of pDNA is desirable.Citation54 Therefore condensing agents such as PEI and Lipofectamine have been used to protect the pDNA during fabrication of polymer loaded films.Citation55 Here, complexation with RALA peptide was able to protect pDNA from degradation within the PVP formulations, but unable to conserve the conformation during the formulation process (). Similar issues with pDNA integrity within coated and dissolvable matrices have been observed previously, although in some cases it has been reported that a loss of pDNA conformation is not necessarily an indicator of biological function.Citation13,18
Indeed transfection studies revealed that the conformational changes in pDNA did not have a detrimental effect on gene expression following release from PVA matrices (). RALA was able to significantly enhance gene expression in vitro approximately 10-fold and thus compensated for a restricted loading capacity within microneedles. Transfection efficacies of pDNA released from PVP matrices were lower, which could be explained by the loss of the supercoiled DNA conformation which was more apparent in PVP matrices, and in particular the PVP-360 formulation, where virtually none of the supercoiled conformation was retained. These transfection rates following release from PVP matrices were similar to those reported by McCaffrey et al (2016) where the transfection efficacy of RALA/pEGFP-N1 (N:P 10) nanoparticles following release from 360 kDa PVP matrices was ∼13% in the NCTC 929 cell line.Citation29 Incorporation of RALA/pEGFP-N1 nanoparticles into PVA matrices improved transfection efficacies to ∼40% at the same N:P ratio (N:P 10). These studies demonstrate a clear rationale for the inclusion of delivery vectors such as RALA in microneedle-assisted DNA delivery systems. Of note is that freshly prepared RALA/pEGFP-N1 nanoparticles (N:P 10) have previously been demonstrated to possess transfection rates as high as 60% in the same cell line,Citation36 indicating that the formulation process could be further optimised to improve pDNA stability within the dissolvable matrix. Similar issues with pDNA integrity within coated and dissolvable matrices have been observed previouslyCitation13,18 and so the addition of stabilisers may be necessary to further conserve pDNA integrity.
Further to these studies it was necessary to determine how much cargo could be incorporated into the dissolvable microneedle arrays. Dissolvable microneedle systems have a limited carrying capacity as the incorporation of large amounts of cargo results in decreased mechanical needle properties.Citation56 In order to circumvent this issue the concentration of polymer solution used to create cargo-microneedles was kept constant (20% or 30% w/w) by diluting highly concentrated polymer stocks with RALA/pDNA solutions prior to microneedle fabrication. RALA/pDNA loaded microneedle retained sharpness and did not lose any of their mechanical robustness. In fact loading of pDNA or RALA/pDNA into the needles actually reduced deformation. This trend was also found by McCaffrey et al (2016) who noted that incorporation of pDNA or RALA/pDNA into PVP microneedle tips did not result in any significant difference to microneedle strength.Citation29 As such, pDNA loaded and RALA/pDNA loaded needles were considered suitable for in vivo applications.
When scaling up for in vivo experiments a large portion of the cargo should be present in the tips and baseplate of microneedles, and should not be lost during the fabrication process. Therefore, following fabrication of RALA/pDNA loaded microneedle arrays, the amount of pDNA residing in the baseplate and needles of arrays was quantified separately from the microneedle sidewalls. The quantity of pDNA present in the baseplate and microneedles varied from 39.4% to 55.0% of the loaded pDNA quantity, and as much as 45% of the pDNA payload was lost to the microneedle sidewalls. Additionally, up to 17% of the pDNA load was not recovered from formulation sidewalls or arrays, indicating a loss of pDNA cargo during the manufacturing process. These limitations could be overcome if microneedles were produced in a large-scale process which localized pDNA only to the tips and baseplate of dissolvable arrays. For example, McGrath et al (2014) described a novel atomised spray process to fabricate dissolvable microneedle arrays from multiple polymer and sugar formulations and were able to restrict the cargo exclusively to the microneedle tips.Citation27 Quantifying the amount of pDNA that could be delivered in vivo was determined via application of RALA/pDNA-loaded microneedle arrays to mouse ears for 24 h. Using this formulation we were able to load and deliver larger quantities of pDNA than other dissolvable devices previously reported, without compromising mechanical integrity.Citation34,35,53,54 Of our formulations, 9–10 kDa PVA microneedle arrays were found to contain significantly higher amounts of pDNA in the baseplate and needles than 360 kDa PVP, 58 kDa PVP, and 13–23 kDa microneedle arrays, and subsequently delivered the greatest quantity of pDNA in vivo. Therefore the evidence from this study indicates that 9–10kDa PVA was the optimal polymer for fabrication of RALA/pDNA-loaded microneedle arrays.
In conclusion this report highlights the importance of careful selection and thorough investigation of the dissolvable microneedle matrix prior to in vivo use, as polymer choice has a significant impact on cargo integrity and loading. These parameters will subsequently impact upon device functionality particularly for vaccine applications where both quantity and integrity of DNA will be critical to generate a potent immune response.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This research was funded by Prostate Cancer UK (S012-006).
References
- Khan KH. DNA vaccines: roles against diseases. Germs 2013; 3(1):26-35
- Cole G, McCaffrey J, Ali AA, McCarthy HO. DNA vaccination for prostate cancer: key concepts and considerations. Cancer Nanotechnol 2015; 6(1):2; PMID:26161151; http://dx.doi.org/10.1186/s12645-015-0010-5
- Saade F, Petrovsky N. Technologies for enhanced efficacy of DNA vaccines. Expert Rev Vaccines 2012; 11(2):189-209; PMID:22309668; http://dx.doi.org/10.1586/erv.11.188
- Sardesai NY, Weiner DB. Electroporation delivery of DNA vaccines: prospects for success. Curr Opin Immunol 2011; 23(3):421-9; PMID:21530212; http://dx.doi.org/10.1016/j.coi.2011.03.008
- Jones S, Evans K, McElwaine-Johnn H, Sharpe M, Oxford J, Lambkin-Williams R, Mant T, Nolan A, Zambon M, Ellis J, et al. DNA vaccination protects against an influenza challenge in a double-blind randomised placebo-controlled phase 1b clinical trial. Vaccine 2009; 27(18):2506-12; PMID:19368793; http://dx.doi.org/10.1016/j.vaccine.2009.02.061
- Daud AI, DeConti RC, Andrews S, Urbas P, Riker AI, Sondak VK, Munster PN, Sullivan DM, Ugen KE, Messina JL, et al. Phase I trial of interleukin-12 plasmid electroporation in patients with metastatic melanoma. J Clin Oncol 2008; 26(36):5896-903; PMID:19029422; http://dx.doi.org/10.1200/JCO.2007.13.9048
- Jackson LA, Austin G, Chen RT, Stout R, DeStefano F, Gorse GJ, Newman FK, Yu O, Weniger BG, Vaccine Safety Datalink Study Group. Safety and immunogenicity of varying dosages of trivalent inactivated influenza vaccine administered by needle-free jet injectors. Vaccine 2001; 19(32):4703-9; PMID:11535320; http://dx.doi.org/10.1016/S0264-410X(01)00225-0
- McCarthy HO, Wang Y, Mangipudi SS, Hatefi A. Advances with the use of bio-inspired vectors towards creation of artificial viruses. Expert Opin Drug Deliv 2010; 7(4):497-512; PMID:20151849; http://dx.doi.org/10.1517/17425240903579989
- Loughran SP, McCrudden CM, McCarthy HO. Designer peptide delivery systems for gene therapy. Eur J Nanomedicine 2015; 7(2):85-96; http://dx.doi.org/10.1515/ejnm-2014-0037
- Lv H, Zhang S, Wang B, Cui S, Yan J. Toxicity of cationic lipids and cationic polymers in gene delivery. J Control Release 2006; 114(1):100-9; PMID:16831482; http://dx.doi.org/10.1016/j.jconrel.2006.04.014
- Mann ER. Review: Skin and the Immune System. J Clin Exp Dermatol Res 2014; 4:3; http://dx.doi.org/10.4172/2155-9554.S2-003
- Venus M, Waterman J, McNab I. Basic physiology of the skin. Surg. 2011; 29(10):471-4
- Stoecklinger A, Eticha TD, Mesdaghi M, Kissenpfennig A, Malissen B, Thalhamer J, Hammerl P. Langerin+ dermal dendritic cells are critical for CD8+ T cell activation and IgH γ−1 class switching in response to gene gun vaccines. J. Immunol. 2011; 186(3):1377-83; PMID:21187444; http://dx.doi.org/10.4049/jimmunol.1002557
- Stoitzner P, Zanella M, Ortner U, Lukas M, Tagwerker A, Janke K, Lutz M.B, Schuler G, Echtenacher B, Ryffel B, et al. Migration of langerhans cells and dermal dendritic cells in skin organ cultures: augmentation by TNF-alpha and IL-1beta. J Leukoc Biol 1999; 66(3):462-70; PMID:10496317
- Valladeau J, Saeland S. Cutaneous dendritic cells. Semin Immunol 2005; 17(4):273-83; PMID:15953735; http://dx.doi.org/10.1016/j.smim.2005.05.009
- Palucka K, Banchereau J. Cancer immunotherapy via dendritic cells. Nat Rev Cancer 2012; 12(4):265-77; PMID:22437871; http://dx.doi.org/10.1038/nrc3258
- Broere F, Apasov SG, Sitkovsky MV, Van Eden W. A2 T cell subsets and T cell-mediated immunity. In FP Nijkamp, MJ Parnham, eds., Principles of Immunopharmacology. Basel: Birkhäuser Basel; 2011; 15-27; http://dx.doi.org/10.1007/978-3-0346-0136-8_2
- McCaffrey J, Donnelly RF, McCarthy HO. Microneedles: an innovative platform for gene delivery. Drug Deliv Transl Res 2015; 5(4):424-37; PMID:26122168; http://dx.doi.org/10.1007/s13346-015-0243-1
- van der Maaden K, Jiskoot W, Bouwstra J. Microneedle technologies for (trans)dermal drug and vaccine delivery. J Control Release 2012; 161(2):645-55; PMID:22342643; http://dx.doi.org/10.1016/j.jconrel.2012.01.042
- Qiu Y, Guo L, Zhang S, Xu B, Gao Y, Hu Y, Hou J, Bai B, Shen H, Mao P. DNA-based vaccination against hepatitis B virus using dissolving microneedle arrays adjuvanted by cationic liposomes and CpG ODN. Drug Deliv 2015; 23(7):2391-8
- Kim YC, Song JM, Lipatov AS, Choi SO, Lee JW, Donis RO, Compans RW, Kang SM, Prausnitz MR. Increased immunogenicity of avian influenza DNA vaccine delivered to the skin using a microneedle patch. Eur J Pharm Biopharm 2012; 81(2):239-47; PMID:22504442; http://dx.doi.org/10.1016/j.ejpb.2012.03.010
- Gill HS, Söderholm J, Prausnitz MR, Sällberg M. Cutaneous vaccination using microneedles coated with hepatitis C DNA vaccine. Gene Ther 2010; 17(6):811-4
- Hu Y, Xu B, Xu J, Shou D, Liu E, Gao J, Liang W, Huang Y. Microneedle-assisted dendritic cell-targeted nanoparticles for transcutaneous DNA immunization. Polym Chem 2015; 6(3):373-9; http://dx.doi.org/10.1039/C4PY01394H
- Mikszta JA, Alarcon JB, Brittingham JM, Sutter DE, Pettis RJ, Harvey NG. Improved genetic immunization via micromechanical disruption of skin-barrier function and targeted epidermal delivery. Nat Med 2002; 8(4):415-9; PMID:11927950; http://dx.doi.org/10.1038/nm0402-415
- Chabri F, Bouris K, Jones T, Barrow D, Hann A, Allender C, Brain K, Birchall J. Microfabricated silicon microneedles for nonviral cutaneous gene delivery. Br J Dermatol. 2004; 150(5):869-77
- Pearton M, Saller V, Coulman SA, Gateley C, Anstey AV, Zarnitsyn V, Birchall JC. Microneedle delivery of plasmid DNA to living human skin: Formulation coating, skin insertion and gene expression. J Control Release 2012; 160(3):561-9; PMID:22516089; http://dx.doi.org/10.1016/j.jconrel.2012.04.005
- McGrath MG, Vucen S, Vrdoljak A, Kelly A, O'Mahony C, Crean AM, Moore A. Production of dissolvable microneedles using an atomised spray process: effect of microneedle composition on skin penetration. Eur J Pharm Biopharm 2014; 86(2):200-11; PMID:23727511; http://dx.doi.org/10.1016/j.ejpb.2013.04.023
- Lutton REM, Larrañeta E, Kearney MC, Boyd P, Woolfson AD, Donnelly RF. A novel scalable manufacturing process for the production of hydrogel-forming microneedle arrays. Int J Pharm 2015; 494(1):417-29; PMID:26302858; http://dx.doi.org/10.1016/j.ijpharm.2015.08.049
- McCaffrey J, McCrudden CM, Ali AA, Massey AS, McBride JW, McCrudden MTC, Vicente-Perez EM, Coulter JA, Robson T, Donnelly RF, et al. Transcending epithelial and intracellular biological barriers; a prototype DNA delivery device. J Control Release 2016; 226:238-47; PMID:26883753; http://dx.doi.org/10.1016/j.jconrel.2016.02.023
- González-González E, Kim YC, Speaker TJ, Hickerson RP, Spitler R, Birchall JC, Lara MF, Hu RH, Liang Y, Kirkiles-Smith N, et al. Visualization of plasmid delivery to keratinocytes in mouse and human epidermis. Sci Rep 2011; 1:158
- Bachy V, Hervouet C, Becker PD, Chorro L, Carlin LM, Herath S, Papagatsias T, Barbaroux JB, Oh SJ, Benlahrech A, et al. Langerin negative dendritic cells promote potent CD8+ T-cell priming by skin delivery of live adenovirus vaccine microneedle arrays. Proc Natl Acad Sci U S A. 2013; 110(8):3041-6; PMID:23386724; http://dx.doi.org/10.1073/pnas.1214449110
- Hirobe S, Azukizawa H, Hanafusa T, Matsuo K, Quan YS, Kamiyama F, Katayama I, Okada N, Nakagawa S. Clinical study and stability assessment of a novel transcutaneous influenza vaccination using a dissolving microneedle patch. Biomaterials 2015; 57:50-8; PMID:25913250; http://dx.doi.org/10.1016/j.biomaterials.2015.04.007
- Sullivan SP, Koutsonanos DG, Del Pilar Martin M, Lee JW, Zarnitsyn V, Choi SO, Murthy N, Compans RW, Skountzou I, et al. Dissolving polymer microneedle patches for influenza vaccination. Nat Med 2010; 16(8):915-20; PMID:20639891; http://dx.doi.org/10.1038/nm.2182
- Kommareddy S, Baudner BC, Oh S, Kwon S, Singh M, O'hagan DT. Dissolvable microneedle patches for the delivery of cell-culture-derived influenza vaccine antigens. J Pharm Sci 2012; 101(3):1021-7
- Mumper RJ, Wang J, Klakamp SL, Nitta H, Anwer K, Tagliaferri F, Rolland AP. Protective interactive noncondensing (PINC) polymers for enhanced plasmid distribution and expression in rat skeletal muscle. J Control Release 1998; 52(1–2):191-203
- McCarthy HO, McCaffrey J, McCrudden CM, Zholobenko A, Ali AA, McBride JW, Massey AS, Pentlavalli S, Chen KH, Cole G, et al. Development and characterization of self-assembling nanoparticles using a bio-inspired amphipathic peptide for gene delivery. J Control Release 2014; 189:141-9; PMID:24995949; http://dx.doi.org/10.1016/j.jconrel.2014.06.048
- Donnelly RF, Majithiya R, Singh TRR, Morrow DIJ, Garland MJ, Demir YK, Migalska K, Ryan E, Gillen D, Scott CJ, et al. Design, optimization and characterisation of polymeric microneedle arrays prepared by a novel laser-based micromoulding technique. Pharm Res 2011; 28(1):41-57; PMID:20490627; http://dx.doi.org/10.1007/s11095-010-0169-8
- Jain AK, Yusuf H, Pattani A, McCarthy HO, McDonald DM, Kett VL. Development of a method to quantify the DNA content in cationic peptide–DNA nanoparticles. J Pharm Biomed Anal 2014; 100:236-242
- Krampe B, Al-Rubeai M. Cell death in mammalian cell culture: molecular mechanisms and cell line engineering strategies. Cytotechnology 2010; 62(3):175-88; PMID:20502964; http://dx.doi.org/10.1007/s10616-010-9274-0
- Larrañeta E, Moore J, Vicente-Pérez EM, González-Vázquez P, Lutton R, Woolfson AD, Donnelly RF. A proposed model membrane and test method for microneedle insertion studies. Int J Pharm 2014; 472(1–2):65-73; http://dx.doi.org/10.1016/j.ijpharm.2014.06.020
- Arda O, Göksügür N. Basic histological structure and functions of facial skin. Clin Dermatol 2014; 32(1):3-13; http://dx.doi.org/10.1016/j.clindermatol.2013.05.021
- Römgens AM, Bader DL, Bouwstra JA, Baaijens FPT, Oomens CWJ. Monitoring the penetration process of single microneedles with varying tip diameters. J Mech Behav Biomed Mater 2014; 40:397-405; PMID:25305633; http://dx.doi.org/10.1016/j.jmbbm.2014.09.015
- Chu LY, Choi SO, Prausnitz MR. Fabrication of dissolving polymer microneedles for controlled drug encapsulation and delivery: Bubble and pedestal microneedle designs. J Pharm Sci 2010; 99(10):4228-38; PMID:20737630; http://dx.doi.org/10.1002/jps.22140
- Lee JW, Park JH, Prausnitz MR. Dissolving microneedles for transdermal drug delivery. Biomaterials 2008; 29(13):2113-24
- Hespe W, Meier AM, Blankwater YJ. Excretion and distribution studies in rats with two forms of 14carbon-labelled polyvinylpyrrolidone with a relatively low mean molecular weight after intravenous administration. Arzneimittelforschung. 1977; 27(6):1158-62; PMID:578432
- Hulme B, Dykes PW, Appleyard I, Arkell DW. Retention and storage sites of radioactive polyvinylpyrrolidone. J Nucl Med 1968; 9(7):389-93; PMID:5713814
- Nair B. Final report on the safety assessment of polyvinylpyrrolidone (PVP). Int J Toxicol 1998; 17(4 Suppl):95-130; http://dx.doi.org/10.1177/109158189801700408
- Morgan AM. Localized reactions to injected therapeutic materials. Part 2. Surgical agents. J Cutan Pathol 1995; 22(4):289-303; PMID:7499568; http://dx.doi.org/10.1111/j.1600-0560.1995.tb01411.x
- Yamaoka T, Tabata Y, Ikada Y. Comparison of body distribution of poly(vinyl alcohol) with other water-soluble polymers after intravenous administration. J Pharm Pharmacol 1995; 47(6):479-86; PMID:7674130; http://dx.doi.org/10.1111/j.2042-7158.1995.tb05835.x
- Kaneo Y, Hashihama S, Kakinoki A, Tanaka T, Nakano T, Ikeda Y. Pharmacokinetics and biodisposition of poly(vinyl alcohol) in rats and mice. Drug Metab Pharmacokinet. 2005; 20(6):435-42; PMID:16415529; http://dx.doi.org/10.2133/dmpk.20.435
- McCrudden MTC, Alkilani AZ, McCrudden CM, McAlister E, McCarthy HO, Woolfson AD, Donnelly RF. Design and physicochemical characterisation of novel dissolving polymeric microneedle arrays for transdermal delivery of high dose, low molecular weight drugs. J Control Release 2014; 180:71-80; PMID:24556420; http://dx.doi.org/10.1016/j.jconrel.2014.02.007
- qiu Y, Guo L, Mao P, Gao Y. Dissolving microneedle arrays for intradermal immunization of Hepatitis B virus DNA vaccine. Procedia Vaccinol 2015; 9:24-30; http://dx.doi.org/10.1016/j.provac.2015.05.004
- Song M, Zeng L, Hong X, Meng Z, Yin J, Wang H, Liang Y, Jiang G. Polyvinyl pyrrolidone promotes DNA cleavage by a ROS-independent and depurination mechanism. Environ Sci Technol 2013; 47(6):2886-91
- Ramgopal Y, Mondal D, Venkatraman SS, Godbey WT. Sustained release of complexed and naked DNA from polymer films. J Biomed Mater Res B Appl Biomater 2008; 85(2):496-503
- Cohen-Sacks H, Elazar V, Gao J, Golomb A, Adwan H, Korchov N, Levy RJ, Berger MR, Golomb G. Delivery and expression of pDNA embedded in collagen matrices. J Control Release 2004; 95(2):309-20
- Vrdoljak A. Review of recent literature on microneedle vaccine delivery technologies. Vaccine Dev Ther 2013; 3:47; http://dx.doi.org/10.2147/VDT.S34682
