ABSTRACT
Quercetin-4′-O-glucoside is one of the major quercetin derivatives in the mature red onion bulb. It has an adjuvant effect on allergies, asthma, arthritis, and cancer. The present study aimed to use uridine diphosphate glycosyltransferase 88A1 (UGT88A1) from Arabidopsis thaliana to achieve the enzymatic synthesis of quercetin-4′-O-glucoside from quercetin. The results showed that UGT88A1 was most active at pH 9.0. The optimum temperature of UGT88A1 for synthesizing quercetin-4′-O-glucoside was 45°C, which was a little lower than that for synthesizing quercetin-3-O-glucoside (50°C). One mutant, V18R, of UGT88A1 was obtained by site-directed mutation and showed a greater affinity (Km 0.20 mM) and twice the enzyme activity (552.3 mU/mg) towards quercetin compared with the wild-type enzyme (0.36 mM and 227.6 mU/mg, respectively). The possible reason could be attributed to the distance change between the 18th amino-acid residue of UGT88A1 and the substrate quercetin, as deduced by molecular simulation.
Introduction
Onion (Allium cepa) and related species have been widely used as flavoring vegetables, folk medicines, and so on, because of their wide range of health benefits, including antioxidant, antiplatelet, antithrombotic, antiasthmatic, and antibiotic effects [Citation1]. In onion, flavonols have been identified as the most important bioactive components. To date, 16 different flavonols have been identified from onion, consisting of glycosylated derivatives of quercetin, isorhamnetin, and kaempferol. Among these, quercetin and quercetin glycosylated derivatives have been found to show a pronounced effect on allergies, asthma, arthritis, and cancers [Citation2]. Quercetin-3,4′-O-diglucoside and quercetin-4′-O-monoglucoside are the major quercetin glycosylated derivatives of the mature red onion bulb (consisting of approximately 93% of the total flavonol content), contributing to the high antioxidant activity [Citation3]. Given the number of secondary metabolites in bulb extracts to be removed in order to purify individual quercetin glycosides, and the notoriously complicated chemical synthesis of quercetin glycosides, it has proven difficult to obtain significant amounts of specific quercetin glycosides [Citation4]. Instead, biocatalysis can be an alternative strategy by which to overcome the limitations of natural product extraction and chemical synthesis for the production of quercetin glycosylated derivatives via glycosylation of the core aglycone molecule. In particular, the regioselectivity of glycosyltransferases (GTs) can efficiently convert quercetin to quercetin glycosylated derivatives, greatly expanding the chemical diversity, functionality, and bioactivity of the core aglycone molecules [Citation5].
Of the 106 families of GTs (http://www.cazy.org/GT1.html), the GTs from Family 1 primarily act on low-molecular-weight substrates with high regioselectivity [Citation6]. In particular, the uridine diphosphate (UDP)-sugar-dependent glycosyltransferases (UGTs) in Family 1 have attracted considerable attention with respect to pharmaceutical and biosynthesis applications due to their unique glycosylation activity [Citation7]. As early as 2003, two onion-derived flavonoid glucosyltransferases were reported, where the regiospecificity of UGT73G1 for the C-3, C-7, and C-4′ hydroxyl groups of the flavan backbone to yield flavonoid mono- and diglucosides was discovered [Citation8]. Quercetin-3,4′-O-diglucoside was probably generated from quercetin through a two-step glycosylation reaction catalyzed by UGT73G1 [Citation9]. However, so far, besides UGT73G1, no other UGTs from onions have been reported that produce quercetin-4′-O-glucoside. Lim et al. found that UGTs UGT88A1 and UGT71B8 from Arabidopsis thaliana, which were classified into group E in Family 1, were able to convert quercetin into quercetin-4′-O-glucoside [Citation4].
In the present study, the codon-optimized gene encoding UGT88A1 was expressed in Escherichia coli, and the enzymatic properties of the recombinant UGT88A1 were determined. The three-dimensional structure model of UGT88A1 and its complex with ligands were built. Combined with analysis of the amino acid sites near the ligand in the model, site-directed mutagenesis was performed to obtain a mutant enzyme with an enhanced activity towards quercetin for quercetin-4′-O-glucoside production.
Materials and methods
Reagents
All the chemicals used were of analytical grade and purchased from Shanghai Aladdin Biochemical Technology Co., Ltd or Sigma-Aldrich Co., Ltd (China). Standards of quercetin-3-O-glucoside and quercetin-4′-O-glucoside were purchased from Tokiwa Phytochemical (Japan, Chiba).
Plasmid and strain construction
The codon-optimized genes encoding UGT88A1 (UniProtKB Accession no.: Q9LK73) from A. thaliana and its mutant V18R were synthesized and inserted into the NdeI–XhoI sites of pRSFDuet-1 with a polyhistidine-tag (His6-tag) at the N-terminus by Genscript Inc. (Nanjing, China). The recombinant plasmids, pRSFDuet-UGT88A1 and pRSFDuet-V18R, were transformed into E. coli BL21 (DE3) to obtain the recombinant strains E. coli BL21 (88A1) and E. coli BL21 (V18R), respectively.
Expression of UGT88A1
The recombinant E. coli strain was cultivated at 37°C in Luria-Bertani liquid medium (10 g/L of tryptone, 5 g/L of yeast extract, and 10 g/L of NaCl) containing 50 μg/mL kanamycin until OD600 reached approximately 0.6. Subsequently, the expression of UGT88A1 and its mutant V18R were induced with 0.1 mM isopropyl-β-d-thiogalactoside (IPTG) at 16°C for 22 h. The cells were harvested via centrifugation at 7822 g for 5 min and the pellet washed twice with 50 mM potassium phosphate buffer (pH 6.8). Then, the cell pellet was resuspended in the same buffer and sonicated using an Ultrasonic Cell Disruptor JY92-IIN (SCIENTZ, Ningbo, China). The supernatant was isolated following centrifugation at 7822 g for 30 min at 4°C. The expression of UGT88A1 was analyzed using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) [Citation10].
Purification of UGT88A1
UGT88A1 and its mutant V18R were purified using a standard Ni-NTA resin (HisTALON Gravity Column Purification Kit; Takara, Dalian, China) according to the manufacturer’s instructions. The purified proteins were concentrated and exchanged into 100 mM potassium phosphate buffer (pH 6.8) using the ultrafiltration tube (Amiconn Ultra-15, Ultracel-50K, Merck Millipore Ltd., Ireland, Cork). The molecular weight and protein purity were determined by 12% SDS-PAGE. The protein concentrations were determined using the Bradford method [Citation11].
GT activity assay
The GT activity assay was conducted as described in a previous study [Citation9]. One unit (U) of GT activity was defined as the amount of enzyme that produced 1 μmol quercetin-3-O-glucoside or quercetin-4′-O-glucoside per minute under the conditions described. The relative activity (%) was calculated using the maximal activity as a control (100%). The effects of pH and temperature on the activity of UGT88A1 were investigated by performing the reactions within a pH range of 4.0–10.0 or within a temperature range of 20–60°C using quercetin and UDP-glucose as substrates. The buffers used were citric acid–sodium citrate (pH 4.0–6.0), potassium dihydrogen phosphate–sodium hydroxide (pH 7.0–8.0), and boric acid–potassium chloride–sodium hydroxide (pH 9.0–10.0).
Kinetic studies of purified GT
The kinetic parameters Km, Vmax, and Kcat were calculated using quercetin (0.5 mM) as a substrate for the enzyme (0.05–0.08 mg/mL). The amount (nM) of quercetin-4′-O-glucoside released by the reaction between substrates and enzyme per unit time was considered as the velocity of the reaction. The kinetic study was performed at 37°C and pH 6.8. The Michaelis–Menten plot was drawn to calculate the kinetic constant. The turnover number Kcat (Vmax/ET) and the catalytic efficiency of purified GT (Kcat/Km) were calculated to determine the GT efficiency.
High-performance liquid chromatography analysis
The sample analysis was conducted on an UltiMate 3000 high-performance liquid chromatography (HPLC) System (Dionex China Limited, Beijing, China) using an Agilent TC-C18 column (250 × 4.6 mm2; America, Santa Clara) maintained at 40°C with ultraviolet detection at 350 nm. The mobile phase A was 0.1% trifluoroacetic acid (TFA) in water, and the mobile phase B was 0.1% TFA in acetonitrile. The flow rate was 1.0 mL/min, the injection volume was 10 µL, and the gradient was as follows: 0.00 min (90% A, 10% B), 8.00 min (75% A, 25% B), 8.01–14.00 min (75% A, 25% B), 14.01 min (75% A, 25% B), 24.00 min (15% A, 85% B), 29.00 min (90% A, 10% B), and 29.01–31.00 min (90% A, 10% B).
Molecular simulation
Multiple alignment of amino acid sequences (UniProtKB accession numbers of which are listed in Table S1) was performed using Clustal Omega (https://www.ebi.ac.uk/Tools/services/web_clustalo/toolform.ebi) [Citation12]. The amino acid sequence of UGT88A1 in the FASTA format was used for model building by SWISS-MODEL (https://www.swiss-model.expasy.org/) [Citation13]. Then, the model of UGT88A1 obtained was uploaded to PROCHECK (http://servicesn.mbi.ucla.edu/PROCHECK/) in the Protein Data Bank (PDB) format [Citation14] to evaluate its reliability. Structures of quercetin and UDP-glucose were obtained from the PDB database, which were used for molecular docking with the modeled UGT88A1 using Autodock 4.2 [Citation15]. For the receptor, a grid box of 80 × 80 × 80 points was defined with a default resolution of 0.295 Å, and the center of the box was set to the geometric center of the bound ligand [Citation16].
The three-dimensional structure model was protonated by means of the H++ server (http://biophysics.cs.vt.edu/H±±) [Citation17] to generate a suitable protonated status for molecular dynamics (MD) simulation. AMBER 18 was used to study the MD of the optimized complex model. The protein–ligand complex was placed in a TIP3P box of size 12.0 Å [Citation18] with force fields 99ffSB [Citation19] and GAFF [Citation20]. The total charge of the system was neutralized with 11 sodium ions (Na+) before energy minimization. The system was equilibrated at a temperature of 303.15 K for 100 ps. Then, the equilibrated complex was subjected to the MD simulation of 1 ns. The total binding free energies of all systems were calculated using the MM/PBSA technique [Citation21] to measure the binding affinity between substrates and a target protein.
Results and discussion
Expression and purification of UGT88A1
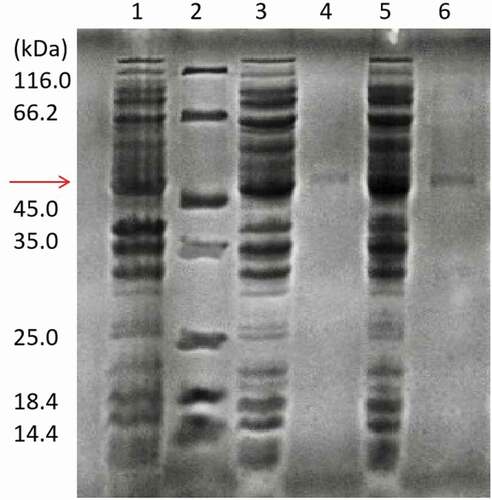
Many plant-derived enzymes are prone to form insoluble inclusion bodies when UGT88A1 is overexpressed in E. coli [Citation22]. In the present study, this challenge was overcome by performing the induction at a lower temperature (16°C) and 0.1 mM IPTG (Fig. S1). The results showed that the soluble expression of UGT88A1 was effective with an obvious band (51.3 kDa) in . Purified UGT88A1 was achieved using the Ni-NTA resin. The activities of purified UGT88A1 for quercetin-3-O-glucoside and quercetin-4′-O-glucoside were 177.6 mU/mg and 227.6 mU/mg, respectively. Figure S2 shows the HPLC chromatogram of catalysis by purified UGT88A1.
Figure 1. SDS-PAGE analysis of recombinant wild-type UGT88A1 and mutant V18R preparations. Lane 1: crude extract obtained from E. coli BL21(DE3) without plasmid; Lane 2: standard molecular marker; Lane 3: crude extract obtained from the recombinant strain E. coli BL21(88A1) with induction; Lane 4: purified wild-type UGT88A1; Lane 5: crude extract obtained from the recombinant strain E. coli BL21(V18R) with induction; Lane 6: purified mutant V18R.
pH dependence of UGT88A1
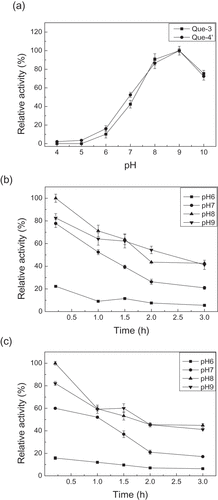
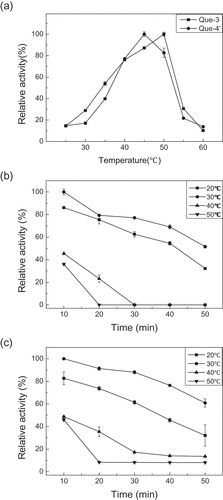
The optimum pH is a critical parameter for an enzyme. The optimum pH of UGT88A1 was found by carrying out the reactions within a pH range of 4.0–10.0. The relative activity (%) was calculated using the maximal activity as a control (100%). ) indicates that the UGT88A1 activity for synthesizing the two quercetin glucosides kept rising from pH 4.0 to 9.0 but the activity was lost under more acidic conditions. However, the activity of UGT88A1 at pH 10.0 was 70% of that at pH 9.0. That is, UGT88A1 preferred a more alkaline environment with an optimum pH of 9.0, which was markedly higher than that of other UGTs, such as UGT74D1 (pH 7.0) [Citation23]. In the study of pH on the stability of UGT88A1, the activity of UGT88A1 declined obviously over time from pH 7.0 to 9.0, but still reserved more than 50% relative activity after 3 h at pH 8.0 and 9.0 (,)).
Temperature dependence of UGT88A1
The temperature pattern of an enzyme is very important for its application. The UGT88A1 reactions were carried out within a temperature range of 25–60°C to determine the optimum temperature. ) indicates that the activity of UGT88A1 achieved the highest value at 45°C (for generating quercetin-4′-O-glucoside) and 50°C (for generating quercetin-3-O-glucoside), whereas higher temperatures inactivated UGT88A1, a finding which was similar to that for other UGTs, such as UGT73G1 (45°C) [Citation9]. In studies on the thermal stability of UGT88A1, the activity of UGT88A1 declined obviously over time when heated from 20°C to 40°C, and only 50% relative activity remained after heating for 1 h at 30°C (,)).
Mutant to improve the enzyme activity
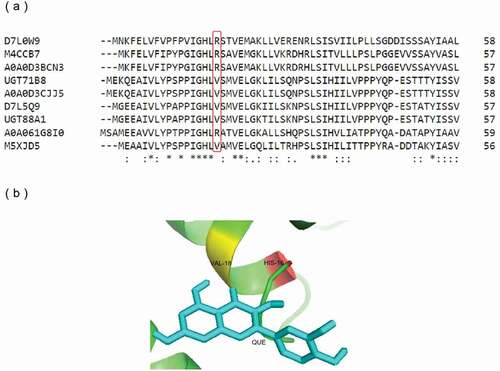
The crystal structure of UGT71G1 (PDB code 2ACV) [Citation24] identified one pocket adjacent to the UDP-binding site, consisting mainly of residues from the N-terminal domain, which would also appear to be the binding pocket for acceptor molecules. Among them, the most important amino acid residue of UGT71G1, His-22, was both close to the hydroxyl group of the acceptor quercetin and the sugar donor. The crystal structure of UGT78G1 (PDB code 3HBJ) [Citation25] indicated the significance of His-15 as the acceptor-binding site. This conserved histidine residue was also observed in the sequence of UGT88A1, that is, His-16.
According to the comparison of multiple amino acid sequences from UniProtKB, of which the Gene Ontology term ‘Molecular function’ included ‘quercetin 4′-O-glucosyltransferase activity,’ the amino acid residues around His-16 in UGT88A1 were relatively conserved, except for Val-18. The amino acid residues at this site could be divided into two categories: nonpolar amino acid (Val) and basic amino acid (Arg) ()). In this study, the mutation of a residue close to the conserved His-16 was considered likely to alter the glucosyltransferase activity of UGT88A1. As a result, Val-18 was chosen to be mutated to Arg-18 (mutant V18R) ()).
Figure 4. Protein sequence and structure analysis of UGT88A1. (a) Part of the multiple alignment of nine UGTs by Clustal O. ‘*’, ‘:’, and ‘.’, respectively, indicate the positions that have a single, fully conserved residue, and the residue with the ‘strong’ or ‘weaker’ groups fully conserved. (b) Stereo diagram of Val-18 and the atoms around (His-16 and quercetin). Quercetin is shown as a stick model.
Enzyme activities and kinetic parameters of UGT88A1 and V18R
On determining the specific enzyme activities (), the mutant enzyme V18R (552.3 mU/mg) showed more than twice the activity for quercetin of that exhibited by UGT88A1 (227.6 mU/mg). The Michaelis–Menten plots of wild-type UGT88A1 and mutant V18R are shown in Figure 3S. The Km value of mutant V18R (0.20 mM) for quercetin was lower than that of UGT88A1 (0.36 mM), showing greater affinity between the mutant enzyme and quercetin. Compared with the Km value of UGT95B2 for quercetin (0.58 mM), which was the UGT newly identified in A. thaliana with a high activity toward flavones and flavonols [Citation26], wild-type UGT88A1 and V18R both had higher affinities for quercetin. Simultaneously, the Kcat value of V18R for quercetin (0.24 s−1) was much higher than that of UGT88A1 (0.16 s−1). Compared with the Kcat value of UGT95B2 for quercetin (0.15 s−1), the Kcat values of wild-type UGT88A1 and mutant V18R were also higher [Citation26].
Table 1. Specific enzyme activities and kinetic parameters of wild-type UGT88A1 and mutant V18R for producing quercetin-4′-O-glucoside.
Assessment of UGT88A1 and mutant V18R
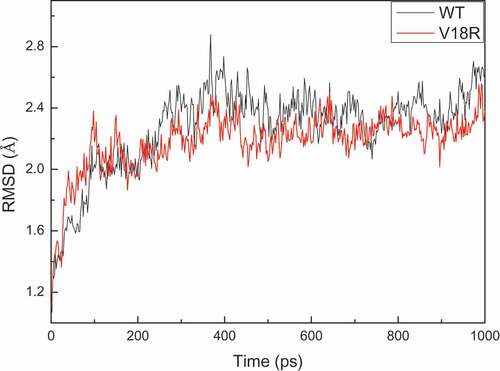
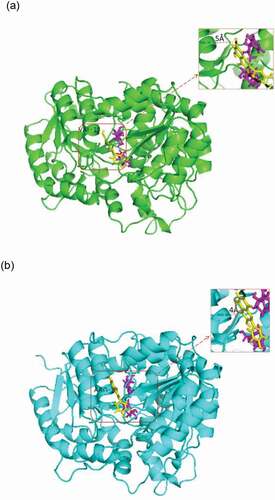
The results showed the better catalytic activity of V18R compared with the parental enzyme UGT88a1 (). A molecular simulation was performed to explain why the mutant V18R affected the activity of UGT88A1. The three-dimensional model of UGT88A1 was first built by SWISS-MODEL. The percentage of residues in the most favored and additionally allowed regions was 99.3%, which showed the reliability of the model (Figure. S4). Overall, the root-mean-square deviation (RMSD) of these two complexes during 1000-ps simulation showed similar conformational stability (). The decrease in free energy after mutation (wild-type UGT88A1: – 21.0620 kcal/mol; mutant V18R: – 27.0474 kcal/mol) further supported the more stable mutant enzyme [Citation27]. Subsequently, the docking results were analyzed, which showed the difference in the distance between molecular and ligand quercetin in the two complex models (). As a result, the distance between the enzyme and the ligand in the V18R mutation reduced from 5 Å to 4 Å after the site-directed mutagenesis, which probably affected the affinity and hence the activity of mutant V18R for quercetin [Citation28,Citation29].
Conclusions
In the present study, glycosyltransferase UGT88A1 from Arabidopsis was expressed in E. coli (DE3) and used to generate quercetin-4′-O-glucoside as the main product in the bioconversion of quercetin. The optimum pH of UGT88A1 was 9.0, and the optimum temperature of UGT88A1 was approximately 45°C. The activity of UGT88A1 was improved by obtaining the mutant V18R, which showed a greater affinity (Km 0.20 mM) and a higher activity (552.3 mU/mg) towards quercetin compared with the original enzyme, probably resulting from the distance change between the 18th amino acid residue of UGT88A1 and the substrate quercetin deduced by molecular simulation.
Compliance with ethical standards
Not applicable
Research involving human participants and/or animals
This study did not involve any experiments on human participants or animals by any of the authors.
Supplemental Material
Download MS Word (211.3 KB)Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Nile SH, Park SW. Total phenolics, antioxidant and xanthine oxidase inhibitory activity of three colored onions (Allium cepa. L.). Front Life Sci. 2013;7(3–4):224–228.
- Ko EY, Nile SH, Sharma K, et al. Effect of different exposed lights on quercetin and quercetin glucoside content in onion (Allium cepa L.). Saudi J Biol Sci. 2015;22:398–403.
- Pérez-Gregorio MR, Regueiro J, González-Barreiro C, et al. Changes in antioxidant flavonoids during freeze-drying of red onions and subsequent storage. Food Control. 2011;22(7):0–1113. . https://doi.org/10.1016/j.jplph.2016.08.017.
- Lim EK, Ashford D. A., Hou B, et al. Arabidopsis glycosyltransferases as biocatalysts in fermentation for regioselective synthesis of diverse quercetin glucosides. Biotechnol Bioeng. 2004;87:623–631.
- Xiao J, Muzashvili TS, Georgiev MI. Advances in the biotechnological glycosylation of valuable flavonoids. Biotechnol Adv. 2014;32(6):1145–1156.
- Lim EK. Plant glycosyltransferases: their potential as novel biocatalysts. Chemistry. 2005;11(19):5486–5494.
- Frederik DB, Jo M, Joeri B, et al. Biotechnological advances in UDP-sugar based glycosylation of small molecules. Biotechnol Adv. 2015;33(2):288–302.
- Kramer CM, Prata RTN, Willits MG, et al. Cloning and regiospecificity studies of two flavonoid glucosyltransferases from Allium cepa. Phytochemistry. 2003;64(6):1069–1076.
- Cai R, Chen C, Li Y, et al. Improved soluble bacterial expression and properties of the recombinant flavonoid glucosyltransferase UGT73G1 from Allium cepa. J Biotechnol. 2017;255:9–15.
- Schägger H, Jagow GV. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379.
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254.
- Chojnacki S, Cowley A, Lee J, et al. Programmatic access to bioinformatics tools from EMBL-EBI update. Nucleic Acids Res. 2017;45:550–553.
- Schwede T, Kopp J, Guex N, et al. Swiss-model: an automated protein homology-modeling server. Bioinformatics. 2006;22:195–201.
- Laskowski RA, Rullmann JAC, MacArthur MW, et al. Thornton, AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol. 1996;8:477–486.
- Morris GM, Huey R, Lindstrom W, et al. Software news and updates autodock4 and autodocktools4: automated docking with selective receptor flexibility. J Comput Chem. 2009;30:2785–2791.
- Marek W. Simplified autodock force field for hydrated binding sites. J Mol Graphics Modell. 2017;78:74–80.
- Anandakrishnan R, Aguilar B, Onufriev AV. H++ 3.0: automating pK prediction and the preparation of biomolecular structures for atomistic molecular modeling and simulation. Nucleic Acids Res. 2012;40:537–541.
- Jorgensen WL, Chandrasekhar J, Madura JD, et al. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926–935.
- Hornak V, Abel R, Okur A, et al. Comparison of multiple amber force fields and development of improved protein backbone parameters. Proteins. 2006;65:712–725.
- Wang J, Wolf RM, Caldwell JW, et al. Development and testing of a general amber force field. J Comput Chem. 2004;25:1157–1174.
- Miller BR, McGee TD, Swails JM, et al. MMPBSA.py: an efficient program for end-state free energy calculations. J Chem Theory Comput. 2012;8:3314–3321.
- Georgiou G, Valax P. Expression of correctly folded proteinsin Escherichia coli. Curr Opin Biotechnol. 1996;7:190–197.
- Jin SH, Ma XM, Han P, et al. UGT74D1 is a novel auxin glycosyltransferase from Arabidopsis thaliana. PLoS Biol. 2013;8:1705.
- Shao H, He X, Achnine L, et al. Crystal structures of a multifunctional triterpene/flavonoid glycosyltransferase from medicago truncatula. Plant Cell. 2005;17:3141–3154.
- Modolo LV, Li L, Pan HY, et al. Crystal structures of glycosyltransferase UGT78G1 reveal the molecular basis for glycosylation and deglycosylation of (iso)flavonoids. J Mol Biol. 2009;392:1292–1302.
- Alexander EW, Sheng W, Li T. PgUGT95B2 preferentially metabolizes flavones/flavonols and has evolved independently from flavone/flavonol UGTs identified in Arabidopsis thaliana. Phytochem. 2019;157:184–193.
- Azam SS, Uddin R, Wadood A. Structure and dynamics of alpha-glucosidase through molecular dynamics simulation studies. J Mol Liq. 2012;174:58–62.
- Singh S, Patel KA, Sonawane PD, et al. Enhanced activity of Withania somnifera family-1 glycosyltransferase (ugt73a16) via mutagenesis. World J Microbiol Biotechnol. 2018;34:150.
- Hans J, Brandt W, Vogt T. Site-directed mutagenesis and protein 3D-homology modelling suggest a catalytic mechanism for UDP-glucose-dependent betanidin 5-O-glucosyltransferase from Dorotheanthus bellidiformis. Plant J. 2004;39:15.