ABSTRACT
The bioengineering tools have significant advantages through less time-consuming and utilized as a promising stage for the production of pharmaceutical bioproducts under the single platform. This review highlighted the advantages and current improvement in the plant, animal and microbial bioengineering tools and outlines feasible approaches by biological and process’s bioengineering levels for advancing the economic feasibility of pharmaceutical’s production. The critical analysis results revealed that system biology and synthetic biology along with advanced bioengineering tools like transcriptome, proteome, metabolome and nano bioengineering tools have shown a promising impact on the development of pharmaceutical’s bioproducts. Tools to overcome and resolve the accompanying encounters of pharmaceutical’s production that include nano bioengineering tools are also discussed. As a summary and prospect, it also gives new insight into the challenges and possible breakthrough of the development of pharmaceutical’s bioproducts through bioengineering tools.
Graphical Abstract
1. Introduction
Most of the organisms have been widely utilized as a potential biofactory of pharmaceutical drugs. Development biology integrated with bioengineering tools for genome manipulation extended the use of several hosts for bioproducts [Citation1]. Biopharmaceuticals are utmost beneficial recombinant bioproteins attained through bioengineering methods. They are resultant from bioresources such as microorganisms, plants, animals or genetically modified tissues and organisms [Citation2]. Bioengineering tools are identically very imperative to expand or upsurge metabolites, which are tangled in their improved endurance and boost their bioactivities [Citation3]. It fastens up new-fangled prospects in the field of pharmaceuticals and biotechnology. Bioengineering tools comprised various procedures for isolation along with the identification of necessary genes from wide-ranging microbes into the host plant genetic material to progress inherently modified or altered host owning innovative features. Though, it drives improved the genetically modified or engineered (GM or GE) crops. It may comprise bioengineering paths for numerous or multiple traits, like secondary complexes, biocatalysts, and proteins [Citation4].
Plant, bacteria, fungi and mammalian cells are the chief biofactories for the production of approximately all beneficial proteins, with a delivery of 68% and 32% in the measure of the global market requirements in 2010 via mammalian and microbial production, respectively [Citation5]. On the other hand, the biopharmaceuticals applications in human healthiness are used for the antitoxin treatment [Citation2]. Currently, many efforts have been in evolution to yield necessary therapeutic complexes, for example, taxol, and artemisinin, etc. shown enormous therapeutic values through metabolic bioengineering tools. Biosynthesis of diverse therapeutic metabolites and various recombinant proteins has been endeavored through the bioengineering tools. Several research data in the literature are presented in this regard [Citation3] such as Mullein by Penicillium janczewskii [Citation6], Taxol by Fusarium maire [Citation7], Aspergillus aculeatinus [Citation8], Pestalotiopsis neglecta and Pestalotiopsis versicolor [Citation9], and Alternaria alternata [Citation10].
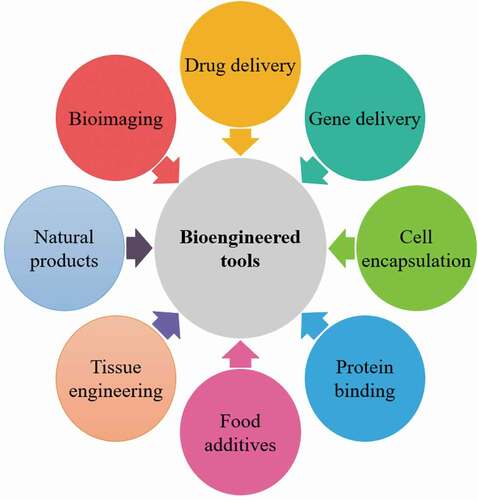
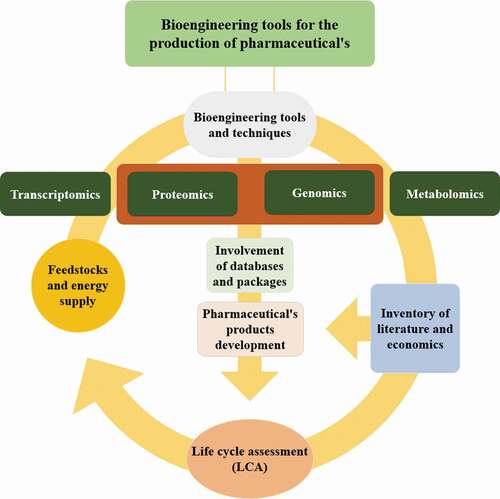
Bioengineering tools are provided genetically modifying organisms for recombinant bioproducts to form medicinal and industrial complexes of high human value. Bioengineering of microbes, animals and plants were widely used for the pharmaceutical product’s production, for example, vaccines, monoclonal antibodies, cytokines, enzymes and growth factors [Citation1]. The existing omics classification included genomics envisioned for DNA, transcriptomics for mRNA, proteomics intended for proteins and peptides, and metabolomics in support of advanced metabolism products. Technologic developments permit parallel investigation of thousands of proteins and genes by high-throughput analysis. Furthermore, hypothesis motivated exploration and breakthrough-driven examination by omics procedures are complementary and comprehensive [Citation11]. Bioengineering tools is a vital part of molecular biotechnology, and genetic engineering. Examples comprise a recombinant bioproducts attained over the fermentation method. Bioengineering for pharmaceutical’s development comprises an inclusive series of techniques and tools (). By this novel comprehensive article, it has been critically analyzed the recent bioengineering tools and techniques through the publication’s on scopus, science direct and other reputed publishers. It could be hastening the progress and implementation of capable bioengineering tools and technologies along with life cycle assessment, which is reported an important pharmaceutical and related problem but not its life cycle assessment. The purposes are to compile these tools and techniques as robust, well-categorized resolutions that achieve an unmet requirement and are capable for enhance the thoughtful knowledge of life science with the bioengineering developments. In this critical review, it is described the current advancement in bioengineering tools for biopharmaceuticals and advancement of the process to obtain industrial important biopharmaceuticals with a global perspective, challenges and future possibilities.
2. Global perspective on pharmaceuticals bioengineered tools
In 1982, the human insulin was the initial recombinant protein through bioengineering tools. It was approved by the FDA for used as a pharmaceutic product. Biopharmaceuticals are revolutionary in the pharmaceutical industry. In accordance with the worldwide profits, 10 biotechnological linked bioproducts are believed among the highest 25 highest selling medicines in 2015; four of them produced by microbes. The higher molecular mass medicines, which can characterize via nucleotides polymers (DNA or RNA) or peptides and proteins (amino acids), are termed biopharmaceuticals [Citation2]. Pharmaceuticals are produced by nucleic acids, for example, siRNA (small interfering RNA), gene therapy and DNA vaccines are very capable pharmaceuticals produced by bioengineering tool’s approaches. Bioengineering tools contain a hill of technologies, extending from outmoded to new pioneering techniques, for example, genomics, proteomics, metabolomics and genetic engineering, which perform a key function in pharmaceuticals sustainability. Pharmaceuticals like monoclonal antibodies are revolutionized the dealing of a variability of illnesses. In current periods, the industrial and academic clusters have started to grow a greater considerate of the natural science of cell lines host, for example, Chinese’s hamster ovary cells and utilize that information for process development and cell line bioengineered tools like ribosome footprint outlining, and stimulating NGS (next-generation sequencing) system that delivers genome extensive evidence on translation [Citation12]. The new-fangled bioengineering tools have given a new stage for enzyme’s bioengineering over the site-specific mutagenesis via a recombinant DNA process to an upsurge the enzyme thermostability. In this respect, the computer-dependent gene designer tools have been established for de novo formation of genetic paradigms. Moreover, gene designer can effortlessly improve, take away, edit, alter and association genetic basics like promoters, tags and open analysis frames [Citation13].
Target proteins have formerly been articulated in plant tissues as screens, but it is the initial report wherein existences of mutant plant tissues expressing an extraneous target protein are used to directly secondary metabolism to a definite pharmacological phenotype [Citation14]. Breitling and Takano [Citation15] have recently studied the new experimental and computational bioengineering tools for the detection, optimization, and manufacture of pharmaceutical biomolecules and outlining the development in the applications and Li et al. [Citation16] examines the bioengineered tools and associated databases for biological parts screening. The growing accessibility of the computerized model building and curation tools additionally upsurges in the accessibility of the bioengineering tools, for example, the wide-ranging computational review of possible production for heterologous paths. Wang et al. [Citation17] described the bioelectronic sensitivity sensor by bioengineered E. coli cells joint with indium tin oxide built electrochemical sensors. Amitai and Sorek [Citation18] have been developed a web-based PanDaTox tool that delivers investigational toxicity evidence for over 1.5 million genes as of hundreds of dissimilar microbial genomes. PanDaTox can fast-track the metabolic engineering developments by permitting the researchers to eliminate toxic genes and confirm the clonability of designated genes before the definite metabolic bioengineering [Citation18].
Nakamae et al. [Citation19] have been described as a newly advanced microhomology intermediated end assembly dependent gene knocks tool, named the PITCh tool, and offered numerous applications. Ventura et al. [Citation20] have described the probiogenomics as a new tool to get genetic perceptions into a variation of probiotic prokaryotes into the human guts. Recently, the genomic sequencing of a huge quantity of Lactobacilli and Bifidobacteria has permitted to access the comprehensive genetic composition of characteristic associates of these bacteria. In the previous two decades, numerous live attenuated and DNA-based vaccines counter to toxoplasmosis are considered, though, only incomplete protection deliberated by vaccination counter to chronic along with acute infection are attained. In this line, Dziadek and Brzostek [Citation21] revealed the recombinant antigen-cocktails was applied as a probable tool for immunoprophylaxis of the toxoplasmosis analysis. The adaptive prokaryotic immune organization CRISPR-Cas is revolutionized the arenas of life sciences and has unlocked the new boundaries to the personalized drug. Kick et al. [Citation22] have been summarized the existing design expansions, novel Cas organizations and its antagonists with current and possible upcoming applications along with the ongoing discussion of ethical problems, which has ascended by the CRISPR-Cas tool.
The CRISPR/Cas9 tools for key genome editing are currently applied for reporting virus resistance in the plants [Citation23]. CRISPR-Cas9 is an RNA directed DNA endonuclease organization, which comprises of Cas9 nuclease showing limited targeting bases through protospacer end-to-end motif complexed by simply customizable sole controller RNA targeting about 20-bp genomic arrangement. They described the various improvements and alternatives of CRISPR-Cas systems, including engineered Cas9 variants, Cas9 homologs, and novel Cas proteins other than Cas9 [Citation24]. To validate the novel luciferase knock-in system, a CRISPR/Cas9 transcription activation/repression system for the PGRN gene was constructed and applied to the knock-in system. In addition, phorbol ester (phorbol 12-myristate, 13-acetate), previously reported as activating the expression of PGRN, was applied to the system [Citation25]. Rojo et al. [Citation26] have also been described that the non-genome editing uses of CRISPR-Cas are also highlighted and a brief overview of the commercialization of CRISPR is provided. Al-Omari at el [Citation27]. have been shown the multi-elemental analysis of pharmaceuticals derived from plant seeds by energy dispersive X-ray fluorescence spectrometry.
3. Significance of bioengineering interventions in pharmaceuticals
Nowadays, bioengineering tools with advanced technologies stipulate variation of the genetic form of plants, animals, and microbes to an upsurge their pharmaceutical’s importance. Metabolic bioengineering could be used as a focus and valuable handling of metabolic paths in a biosystem by employing regulatory and enzymatic roles of the cells via biotechnological involvement like rDNA (Recombinant DNA) technology [Citation3]. Bio-advancement of low price and high-yield procedure is the main goal of bioengineering tools. Hence, the amplified metabolic actions can be accomplished more competently by employing the bioengineering tools and techniques from biotechnology, other recombinant with molecular biology procedures. On the other hand, nano biotechnology is a state-of-the-art cutting-edge area encompassing the bioengineering tools to form pharmaceutically useful nanomaterials (NMs) or a combination of nanocomponent myriad to generate devices/stages through exceptional biological, physical and chemical interface properties [Citation4]. As per the elemental configuration, these could be collected of carbon-based polymers (synthetic/natural), radionuclides and mixtures showing diverse morphologies (rods, elements, tubes, films and lattices), and dimensions proportions. It could be biosynthesized through numerous tops to bottom methods. These may include tools intricate in bioengineering with tissue culture, rDNA technology, including polymerase chain reaction, gene cloning, genomic transformation and transgenics. A recombinant reporter phage (light-tagged) has been generated by mixing the luxAB genes to the P. cannabina phage PBSPCA1 complete genome. The reporter phage is recognized the P. cannabina from diseased specimens’ representative the possible of the diagnostic for identification of disease. The reporter phage shows the capacity for the speedy and exact diagnostic recognition of cultured isolates, and infected specimens [Citation28,Citation29].
Novel bioengineering tool’s breakthroughs are accomplished by rethinking how we see and handle key challenges in the pharmaceutical’s generation. A collective outline of numerous dynamic and progressively advancing ranges that are forming the next era of bioengineering tools pointing to supply more precise cellular intuitive, higher exactness, quicker observing, more specific delivery, and more accurate forecasts for pharmaceutical’s production. All through these distinctive ranges, a common theme is the continuous thrust to make strides accuracy, affectability, and selectivity; which are bringing us closer to the improvement of personalized bioengineering tools.
4. Integration of system biology and synthetic biology into bioengineering
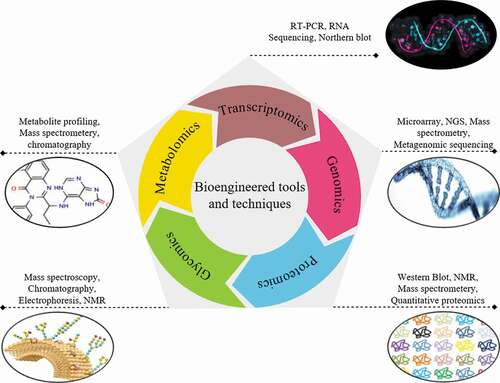
To comprehend the worldwide framework of a metabolic scheme in an improved way and to offer the modern advance approaches of bioengineering strongly, the mandate of complete methods has been enormously amplified. The elementary principle of systems biology is to produce combined multi form data of functional biosynthetic paths in plants, animals and microbes [Citation3]. With current developments in high-throughput sequencing, the swift accumulation in evidence of biological statistics are customary for diverse points, i.e., genomic, proteomic, transcriptomics, metabolomics and fluxomic, etc. required.
4.1. Systems biology for bioengineered strategy
The systems biology encompasses newer advanced methods of considerate high-throughput information recovered from numerous omic methods through computational imitation as shown in [Citation3]. The procedure is repeated until the end of developing better-quality microorganisms with desired qualities is attained. This cyclic repetition includes the utilization of transcriptomic information. Moreover, from RNA-seq over sequencing technology, especially subsequent generation. Sequencing tools or microarray is provided the proteomic information, while LC/MS, GC/MS and NMR produced metabolic summarizing information or metabolome along with labeled isotope’s materials produced information from metabolic flux investigation (MFI). The estimates of the subsequent circular alteration of multiple/solitary genes are then carried forward through assimilating the evidence by bioinformatics for display the omics information. The overview of the high-throughput sequencing tools in consort through the bioengineered phenotyping strategy for phenomics could be improved the investigation of non-prototypical plants through transcriptome; genomics sequencing with computational science [Citation30]. With current developments in high-throughput sequencing tools, the speedy accumulation of biological data statistics set at diverse stages, i.e., transcriptomic, genomic, proteomic, fluxomic, and metabolomic etc. occupied a place. The regulation levels are required for elaborate the evidence about the signal transduction paths along with the transcriptional directive, protein trafficking and many more. Metabolic bioengineering may be the integration of wet laboratory trials (high-throughput experiments) as of in silico procedure. A successful data set has utilized the information produced from all omics connected methods and revealed the potential for upcoming directions [Citation3].
Table 1. Several active databases record for systems biology investigation with their corresponding web links.
For recovering the important information to understand the multiplicity and exclusivity of metabolic paths, the recognitions for organism enhancement with an examination of genomics are a probable and operative method. The genes which are not significant for the desired biochemical mixtures, it could be identified, in addition. These genes accompanied by selected genes could be made known from the start the innovative path’s flux [Citation50]. Currently, the numerous genome missions have been proficient using NGS stages [Citation51]. Genetically reduced trypanosomatid parasites could be attained by the omission of genes. Gene’s deletion shows the benefit that these parasites can undertake homologous recombination between endogenous and external DNA sequences insincerely introduced in the cells. This bioengineering tool has simplified the detection of numerous unknown gene functions, along with permitting us to take risks about the possible for genetically reduced live organisms as investigational immunogens [Citation52].
4.2. Transcriptome based analysis
The transcriptomes study is probable by using next-generation sequencing stages, as a result of the low price associated with the distinctive capability for de novo assemblage and transcriptomic richness quantification. It has been provided improved considerate of varied plant’s species, specifically for therapeutic properties [Citation53]. The extensive identification is probable by employing the data from extremely sequenced transcriptomes produced via next-generation sequencing. For examination, the numerous plans are accessible, which employ locus genomic data. Such packages included recognized and applied software, for example, Bowtie, TopHat and Cufflinks. These bioprograms support in the sequence reads arrangement of the genome and support in the transcript abundances through statistical valuation. At that point, the assemblage bioprograms and quality evaluation system (CONSED, PHRED and PHRAP) are extremely valuable for RNA-seq dependent analysis. On the other hand, the main application of transcriptome assemblage is the manifold k-mer tool. The assemblers applying such tools are Oases and Trans-ABySS [Citation54].
4.3. Proteome-based analysis
The proteome outlining is the good selection which can be achieved by the acquisition of data as of metabolic responses catalyzed through diverse enzymes. Such analysis is started by the protein’s separation through the 2D electrophoresis system. Then, the purified protein is considered through mass spectrometry. Furthermore, by two or extra flexible situations of genotypic upbringings, the relative examination of protein particles through transformed strengths can be observed simply. There are several cases display positively confirmed the enhancement of diverse traits. Proteins are processed deprived of gel separation along with LC-MS could be applied for recognizing the split peptides. The produced peptide arrangements are exposed the record exploration. This process is valuable and fast over the 2D gel examination tools [Citation55]. Therefore, these tools could support proteomics and its character in the systems biology and bioengineering strategy. The tool for the phagosome proteome is a treasured approach to measure the capability of a normal or hereditably engineered prokaryotic strain to moderate the targeted immune response [Citation3]. Protein refolding is a vital process for gaining dynamic recombinant proteins from insertion bodies. Though, the conformist refolding process of dilution or dialysis is time-consuming and improved active protein produces are often small, and a bulky trial with error procedure is essential to attain success. Investigators are used manageable diffusion over laminar airflow in microchannels to control the denaturant attentiveness. This commentary introduces the ethics of the protein refolding technique via ‘microfluidic chips’ and the benefit of results as a bioengineered tool for fast and resourceful recovery of lively recombinant proteins from insertion bodies [Citation56].
4.4. Metabolome-based analysis
Metabolic bioengineering of terpenoids developed as a modern strategy to create actually occurring bioproducts. Plants synthesize the common precursor for isoprenoids, isopentenyl diphosphate, and its allylic isoform dimethylallyl diphosphate by two particular and compartmentalized pathways, the cytosolic mevalonate pathway and the plastidial MEP pathway [Citation57]. The enormous amount of plant metabolite’s structure of insignificant portions is recognized and identified. The main emphasis of metabolomic is the recognition of metabolites and measurable concentration’s resolution. An amount of high-resolution and high-throughput tools has been established for analysis and identification of cellular metabolites showing the metabolic position of cells [Citation58]. Mixtures of procedures are typically ideal due to its improved firmness along with well applicability. GCMS is suitable for the chief metabolite’s examination tool in essential metabolic paths of plants, like sugars, carbon-based acids, amino acids in tricarboxylic acid series and fatty acids, where LCMS is appropriate for phosphate sugars, secondary metabolites and cofactors. Correspondingly, mass imaging system also delivers a capable method via NMR wherein mass signals formed by plant tissues scanned by diverse ionization approaches are composed for the localized metabolites. These tools permit the recognition and quantifiable assessments of a metabolites with operational convolutions and adjacent resemblances [Citation3].
5. Tools for the bioengineering of pharmaceuticals
To achieve countless purposes of systems biology via modeling and simulation, a lot of tools and database packages are technologically advanced. These tools are continuously being developed to simplify the engineering strategies for biopharmaceuticals (). The latest update (July 2019) contains many tools and databases. The user can be able to explore and query the database by entering related terms. These may broadly describe as follows.
5.1. Unified metabolic database and networks
The metabolic data network records comprise necessary evidence related to metabolic routs, bioreactions, and enzymes essential for the description of the physiological features and metabolic of an organism (). BioSilico is an Internet web-based record system that simplifies the exploration and metabolic pathway’s analysis. Heterogeneous metabolic data, including ENZYME, EcoCyc, LIGAND and MetaCyc are combined in an organized way, thus allowing users to resourcefully retrieve the applicable evidence on biochemical compounds, enzymes and reactions [Citation59]. Radrich et al. [Citation60] have applied this procedure for the metabolic system of the Arabidopsis thaliana plant. A systematic assessment of compounds and their reactions between two genomes are extensively permitted databases to acquire a high-quality main consensus rebuilding. GEnome-scale Metabolic Models (GMMs) and BiGG Models are a treasured tool, since they characterize the metabolic assembly of cells and deliver an efficient scaffold for quantifying metabolic fluxes and simulating in the organism’s system through constriction based mathematical approaches. The combination of omics information into the structural data is well defined by GMMs permits to build additional accurate evidence of metabolic positions [Citation61].
Table 2. Bioengineered web-based tools for pharmaceuticals developments.
In the view of significant application of metabolic change analysis in considerate of metabolic bioengineering, it turns out to be vital for user responsive computer-based package. It can provide the excessive support to the investigators with the comprehensive computational approaches. Consequently, a package of MetafluxNet has been established, which offered a significant biology systems stage for metabolic engineering and description [Citation78]. In bioengineering tools, typically there are generally three schemes, which are often used, for example, whole plants, tissue culture cell, and plant genes in microbes [Citation3]. By bioengineering, favorite proteins are formed in large amounts to chance the abundant stresses of bioindustry. Henceforth, the maximum biopharmaceuticals formed nowadays are recombinant [Citation79]. PITCh designer can automatically design not only the appropriate micro homologies but also the primers to construct locus-specific donor vectors for PITCh knock-in. By using the newly established pipelines, a reporter cell line for monitoring endogenous gene expression, and transgenesis (TG) or knock-in/knockout (KIKO) cell line can be produced systematically [Citation19].
5.2. Protein bioengineering tools
Protein bioengineering tools involved alteration of protein sequence at the molecule level with the intention of alter its biofunction. The utmost uses in the protein system are base pair cuts as well as connections. Though, the protein structure changes are triggered by oxidation or unalterable decrease of disulfides are similarly measured. One aspect that paid conclusively to protein bioengineering is the technique’s advance that allowed the fortitude of protein’s three-dimensional assembly at the molecular level. Between these tools, the supplementary prominence is certain to the X-ray crystallography tool as a result of its high tenacity (reaching <1Å). Further newly, Cryo-electron microscopy and nuclear magnetic resonance (NMR) have also grown space as alternate tools for resolving structures [Citation2]. Dwane et al. [Citation80] have described the tools used for the study of protein complexes assembled in signaling cascades.
5.3. Metabolic bioengineering tools
Metabolic bioengineering tools are very vital to recover or upsurge the metabolites which are complicated in plant and microbial improved existence and increase their cellular in addition to biological actions [Citation3]. Metabolic bioengineering is referred to as the focused and beneficial alteration of metabolic system working inside the living cells by enzymatic, transporters and regulatory purposes via the recombinant DNA (rDNA) approach. Metabolic bioengineering can be advanced over up to regulation or overexpression of selected genes, for example, improved tocopherol formation through overexpression gene, called geranylgeranyl transferase. Currently, the existing approaches and devices for altering microbes have been growing, comprising advancement in the design of ribosome binding positions, genetic promoters, riboswitches, modular vector arrangements, reporter proteins and without marker selection organizations. As a result of novel toolkits, microbes have been effectively bioengineered to express diverse heterologous paths for the generation of an extensive variation of treasured bio-compounds. In silico approaches have been active in an upsurge their bioproduction possibility [Citation81].
Medicinal and non-medicinal plants are widely utilized as a chemical biofactories due to the ability to capture the solar energy. The rDNA bioengineering tool is the root of metabolic bioengineering of these plants by which form a transgenic variety of plants. Crop plants are utilized for the mass formation of various plant chemicals, specifically in the pharmaceutical’s field [Citation3]. Currently, tissue culture tools are commonly used practice for gaining the huge number of phytoconstituents in the sort of period. Plant tissue environments can be maintained under control mode (for example, example elicitor’s and stress enhancement action) that the suitable gene displays might be articulated. The advanced stages of metabolites are produced and collected. In tissue culture cell line, the metabolic outlines could be reformed by altering genes, which encode production enzymes or controlling features of the metabolic path of curiosity. Tissue culture can also deliver an exposed area for gene’s screening that could be additional supportive for the formation of a transgenic variety of plants regarding to specific feature [Citation82].
Generation of a transgenic variety of plants proceeds more development periods than the progress of microorganisms. Carotenoid in addition to isoprenoid biosynthetic paths are hereditably changed via bioengineering tools. These bioengineering tools can also be utilized for the functional identification of a transgene encrypting putative enzyme. In such tools, the plant genes can easily be cloned in an appropriate vector and thereafter altered into the host microbes. Construction of transgenic plants takes an additional period than microbes’ culture. The current thrust is that biotechnology’s tools attracted to discovered new enzymes and genes for improved considerate of metabolite’s flux. Some non-traditional recent approaches, by which metabolic flux is amplified, and identified the genes through genome or transcriptome mining, concurrent expression outlining of numerous genes in plants, escapist flux through silencing and overexpression. Bekker et al. [Citation83] have summarized and discussed the published literature on tools for metabolic engineering of Streptomyces over the last decade. These strategies involve precursor engineering, structural and regulatory gene engineering, and the up or downregulation of genes, as well as genome shuffling and the use of genome scale metabolic models [Citation3].
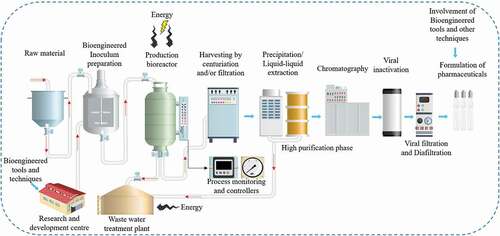
5.4. Bioengineering upstream tools
The key area of the development course is the revolution of growth into the anticipated metabolic bioproducts via bioengineering tools [Citation2]. This involves well-regulated environments and contains the usage of industrial-scale bioreactors tools. Numerous features should be measured by using the bioengineering tools, for example, process nature (batch, continuous, fed batch and temporary immersion), pH, temperature and oxygen source regulator, material’s sterilization and environmental maintenance to confirm it is free from microorganisms. On the other hand, by streamlining upstream forms and utilizing a nonspecific virus‐like particle’s base, virus‐like particles stage bioengineering tool is anticipated to diminish the administrative stack of person immunizations given that controls for the base and its filtration will be ended up well characterized. Virus‐like particle’s stages may indeed quick track immunization conveyance in reaction to widespread circumstances [Citation84].
5.5. Bioengineering downstream tools
Downstream bioengineering tools comprised of all stages essential to separate a bioproducts from growth culture mediums to ultimate purified bioproduct. It contains numerous stages to detention the target biomolecules and to eliminate host tissue connected impurities (for example, example DNA, cell proteins), course associated impurities (for example, example leached ligands, buffers, antifoam) and product associated contamination (e.g., fragments, aggregates, clipped species, etc.). Downstream bioengineering tools typically included three key phases, namely (i) preliminary recovery (taking out or separation), (ii) distillation (elimination of impurities), and (iii) improving (elimination of definite impurities and undesirable types of the specific molecule that might have designed in separation and purification).
Preliminary repossession includes the separation among cell as well as supernatant. If the key molecule is formed extracellularly, the elucidated broth is applied for absorption (such as ultrafiltration) after that purification. Perhaps, the concealed and solvable proteins in the growth medium of P. pastoris could be straight stored by centrifugation. The samples could be concerted, and the significant protein separated from the supernatant by precipitation, ultrafiltration, along with chromatography. For intracellular molecules, the cells collected must be transferred to lysis (sonication, pressured to homogenize, passing over mills) afterward elucidation to eliminate cell debris [Citation2].
6. Nano bioengineering tools
6.1. Nano bioengineering enabled transformation tools
The ultra-very minor dimensions of the nano bioparticles prepared them with the capacity to intrude on the membrane barriers and cell wall deprived of any forcible pass techniques. The interior payload of functional nano-materials in plant tissues may transpire either via passive dispersal over cell partition present in the apoplast area providing the nano-material dimension is lesser than five nanometer by crisscrossing via transmembrane networks, phospholipid bilayer and/or by the dynamic procedure of endocytotic vesicle development. The former investigation on the plant host deals to the biosorption of nanoparticles, as a result of the wide application in bioindustry and easy to detect and tracing by microscopic tools. Though, it likened to the excessive wealth of evidence accessible in metazoans, solitary a few of combined reasonable analyses are considered to establish the involvement of the physicochemical characters of nanoparticles in plant–nanoparticle interface [Citation85]. The major nano-material for distribute the genes which comprised of silica nanoparticles, gold nanoparticles, single-layer carbon nanotubes, multiple-walled carbon nanotubes, calcium phosphate nanoparticles, magnetic nanoparticles, iron oxide particles, zinc oxide with sulfide nanoparticles, cadmium sulfide or quantum dots, functionalized magnesium phyllosilicate, amino clay nanoparticles, polymeric nanoparticles (liposomes, poly-L-lysine, poly-ethyleneimine, dendrimers, cellulose nanoparticles), and many others [Citation4]. Fluorescent nano-diamonds are capable of nanoprobes, because of their constant and magneto-sensitive fluorescence. Hemelaar et al. explained the generally applicable bioengineering tools for the passage of fluorescent nano-diamonds into Saccharomyces cerevisiae. Their cells are a favorable model to examine the intracellular bioengineered processes. They have been assessed electrical transformation along with situations of chemical permeabilization for acceptance of competence and feasibility [Citation86].
6.2. Bioengineered gene gun tools
Bioengineering tools are included a transfer of nucleic acid ballistically by gun precipitate liberation or controlled gas compression tools counting the usage of nitrogen, flattened air and inert helium gas for fast-tracking the tungsten or gold micro-projectile units in plant tissues. Generally, metal or its oxides are applied for biolistics as nonmetal oxides less dense or lighter at nano-scale than the others [Citation4]. On the other hand, the chemical expression inducers are overloaded into mesoporous silica nanoparticles pores (approximate 3 nm) that are then covalently plugged with gold nanoparticles. The capped mesoporous silica nanoparticles are then covered with plasmids and distributed by gene gun to N. tabacum cotyledons, in which protein expression is activated upon unsealing and release of the appearance of inducer [Citation87]. Compatibly, Martin-Ortigosa et al. [Citation88] have strained to enhance the experimental constraints via gold-plated mesoporous silica nanoparticles and gold nanorods, co-bombardment by microparticles and improved DNA-nano particles addon protocol to carry DNA in onion epidermis cells, maize, and tobacco foliage explants [Citation4]. Such tools can also deliver protein over the wide use of gene gun organization. It can be readily useful to any organism or tissue amenable to biolistics tools [Citation88].
6.3. Nanopore genomic sequencing tools
Nanopore sequencing tools have the more potential to concentrate other sequencing tools outdated with its capacity to produce long reads and delivers compactness. The such nanopore tools are of critical importance, as they should overcome the high error rates of the other traditional tools [Citation89]. The compassion of the quarter age sequencing tool is to the close of single nucleoside to be as huge as kilobase measurement of RNA or ssDNA that can be sequenced deprived of the essential for PCR amplification or labeling through radioactive, organic, and fluorescent markers. The Oxford nanopore tool is the MinION long-read sequencer for repetitive whole-genome by genome sequencing of an ataxia-pancytopenia condition and severe immune dysregulation [Citation90]. Moreover, the basecalling tools with advanced accuracy and presentation, like Scrappie, can overcome the key disadvantage of nanopore sequencing tool, i.e. higher error. The overlay bioengineering tools are Minimap and GraphMap, achieve, likewise, in terms of accurateness. However, Minimap achieves improved than GraphMap in terms of rapidity and recollection usage by storage only minimizers in place of all k-mers, and GraphMap. A state-of-the-art polishing bioengineering tool, Racon, produces high-quality consent sequences while provided an important speedup over the additional polishing tool, nanopolish [Citation89]. The ascendable pipeline for the real-time examination of MinION sequence information and use of such as a pipeline to display preliminary proof of notion that metagenomic MinION sequencing bioengineering tool can deliver rapid, precise diagnosis for prosthetic combined infections [Citation91].
6.4. Bioengineering imaging tools
The nanobiotechnological involvements are giving microscopes with great higher resolutions as there are far better-quality nano permitted silicon dependent solid-state assembly tools accessible now to build compressed yet vigorous transportable microscopes. A nanoparticle is typically categorized via spectroscopic and microscopic tools [Citation92]. These can be used as beneficial mediators (magnetic nanoparticles producing hyperthermia), as drug transporters, or as contrast mediators for imaging resolutions. The nanomaterials could be biocompatible, well categorized, and steady in vivo [Citation93]. These nanoparticles have numerous advantages with real-time intensive care, negligible or no insensitivity, user-friendliness without tissue obliteration [Citation94]. Regarding cancer imaging, several investigations have exposed that superparamagnetic iron oxide nanoparticles conjugated to folic acid can be oppressed as difference mediators for magnetic resonance imaging [Citation93]. Photoacoustic imaging tools have developed as a hopeful imaging stage with a high tissue diffusion complexity. The expansion of such a nanoparticle embraces the great potential for clinically adaptable diagnostic imaging with higher biocompatibility. In vivo photoacoustic imaging tool trials in mice have confirmed that the nanoparticles gather in lymph nodes, highlighting their possible usefulness as photoacoustic mediators for sentinel lymph node recognition. The conjugate imaging tools are also multipurpose to distinguish the plants nutrient position. Hyper supernatural imaging produces nondestructive actual time three-dimensional localization of a specific micro or macronutrient in plant tissues feasible. This will transfigure to be considerate on nutrient separating, redistribution and translocation between diverse plant tissues at numerous growing plant phases [Citation95].
6.5. Bioengineering nano-tools for therapy and diagnostic products
Bioengineering nano tools will contract with emerging nano biotechnology-derived cheap products for extremely subtle, speedy, and consistent plant-pathogen discovery to confirm initial illness diagnostics [Citation4]. There are sufficiently reports documented the antifungal, antibacterial and antiviral actions of various kinds of nano materials [Citation96], but the expansion of new theragnostic mediators will be more required [Citation97]. Such nano materials will not only hold the shallow ligands leveling them to spread at a specific position using simplistic paths and plant apoplast but also be encouraged by positive inducements comparable visual. These are thermal, magnetic or acoustic to either extricate the beneficial payload or may pledge to impair properties on the causal agent ‘the pathogen’ or on the diseased tissue, to cause local cell decease and necrosis to restraint the feast of the illness to uninfected materials [Citation97]. Ozkan et al. [Citation98] described the utility of an immunoglobulin Y alkaline phosphatase bioconjugate as an investigative tool for the influenza-A virus. M2e definite IgY antibody is conjugated covalently to the alkaline phosphatase enzyme by the occurrence of glutaraldehyde. The antibody-dependent enzyme bio conjugate is considered by Fourier-transform infrared and fluorescence spectroscopy.
6.6. Bioengineering nano-barcoding tools
The barcodes are rapid elasticities of gene arrangements or RNA, ssDNA or complementary DNA that can be coated or immobilized on the exterior of a nanoparticle or nano materials (nano-barcodes) to perceive specially in numerical form and enumerate target genomic mRNA, biological model complementary DNA, chiefly plant quantifiable [Citation4]. Geiss et al. [Citation99] have established a nano-string counter DNA barcoding visual fluorescence tool to recognize and enumerate straight mRNA from the model deprived of the obligation of extension and development of complementary DNA. A DNA worked silica-coated gold nanoparticle-based nano-barcode assay concerning surface improved Raman’s spectroscopy examination are described to distinguish transgenic Bt (Bacillus thuringiensis) from rice [Citation100]. Prearranged nano-particles have been utilized for barcoding and are in the countless mandate for the real-time analysis of manifold targets. The encoding tools to produce unique manifold codes for nano-barcodes are depended on confident encoding elements together with optical (non-fluorescent and fluorescent), graphical, magnetic and segment change possessions of nanoparticles or their diverse shapes and dimensions [Citation101]. In conjunction with speedy nanoparticle biosynthesis, high-throughput in vivo nanoparticle screening tools will permit researchers to trial more particles instantaneously than ever before, concentrating on DNA barcoded nanoparticles [Citation102].
7. Production of pharmaceuticals through bioengineering
Although some diverse expression organizations may be employed novel technological advancements for mammalian cell lines, plants, insects, which are endlessly being made to recover biopharmaceuticals production from microbes. This outlay is acceptable via the healthy categorized genomes, adaptability of plasmid vectors, obtain the ability of diverse host species, price efficacy as associated with the additional appearance of organizations [Citation1]. De novo, biosynthesis of folates in plants is strongly controlled through response regulation of certain path catalysts. Currently, Chaudhary et al. [Citation103] explored the forecasts of continuous manufacture of folates in a progressive conjunction, through the high expression of targeted response and evolutionarily preserved heterologous bacterial (E. coli) dihydroneopterin aldolase in tobacco. They have defined the biotechnological probable of such allosterically separated trans molecule for enhanced folate manufacture in plants for upcoming folate agri-biofortification outlines.
7.1. Pharmaceutical’s important recombinant protein
A bioengineered model alteration arisen in 1973 with the encounter of a rDNA tool by Cohen and Boyer, and the fruit’s discovery were appeared in 1982, with the governing consent of recombinant human insulin formed by E. coli genetic engineering [Citation104]. Between 100 and 200 health, beneficial protein is accepted in Europe and USA. Non-glycosylated proteins are generally formed in E. coli, and they establish 55% of the beneficial protein global markets. N-glycosylated proteins are typically formed in mammalian cells which imitator of human glycosylation. Chinese hamster ovary cells deliver about 35% of the beneficial protein global industry, but the procedure is highly costly, and the glycoproteins made are not accurately the human nature, and in other cases, they must be altered. Ten percentages of the global industry are provided by supplementary mammalian organizations such as mouse myeloma cells. Insect cells are similarly used. Though, molds, yeasts and insect cells are usually incapable of deliver mammalian glycosylation, the widespread methylotrophic yeast strain, Pichia pastoris, has been inherently planned to yield a human form of glycosylation [Citation79].
A. Bacteria
Due to the rapid developments of bioengineering tools like the recombinant DNA technique, human proteins might be attained by heterologous appearance using E. coli, in addition to bacteria. The typical case is a human insulin. It is utilized to manage diabetes. At first, insulin was separated from the porcine pancreas and bovine extracts [Citation2]. Bacterial organizations are utilized to form somatostatin, bovine growing hormone and insulin for veterinary submissions, α1 antitrypsin, interleukin two, tumor necrosis influence, β-interferon and γ-interferon. An enhanced Gram-negative bacterium for recombinant protein formation has been established using Ralstonia eutropha. The system seems higher to E. coli with reverence to enclosure body formation. Organophosphohydrolase protein is likely to enclosure body development with a manufacture of lower than 100 mg per liter into E. coli, was formed at 10 g l per liter in R. eutropha. Bacteria like S. carnosus could yield 2 g per liter of concealed mammalian protein. The consistent of target-specific gene expression power of the Sec translocon volume could be used to progress toward the yields of a recombinant E. coli protein in the periplasm [Citation105].
B. Yeasts
Yeast has a significant function to play in membrane protein organizational biological research [Citation106]. They are extensively used in the generation of recombinant proteins for industrial and medical interest because they purely and competently meet requirements for both platform and market drugs. There is a wide variety of recombinant protein significant yeast expression species, including Saccharomyces cerevisiae, Hansenula polymorpha, Pichia pastoris, Kluyveromyces lactis, Yarrowia lipolytica, Schizosaccharomyces pombe, and Arxula adeninivorans [Citation107,Citation108]. Furthermore, it has been described that Yarrowia lipolytica is secreted high points of heterologous proteins, for instance, 1–2 g/L alkaline extracellular protease [Citation109]. High recombinant protein yields can be obtained in the methylotrophic yeast, P. pastoris, e.g., as high as 15 g per liter. It was found that P. pastoris can make 20–30 g per liter of recombinant proteins. For such strain, two new promoters have been positively recognized and could probably be functional in recombinant protein expression, such as promoter P0547 (methanol‐inducible) and promoter P0472 [Citation7]. Furthermore, by addressing the translation and transcription of mRNA coding for recombinant protein, substantial yield enhancement has been attained from Pichia pastoris [Citation110]. Furthermore, with developments in robotics technology, the exploitation of real-time yeast hosts in high-throughput screening will convert more regular to regulate the desired yeast recombinant protein.
C. Mammalian cells
Chinese hamster ovary cells establish the favored scheme for manufacturing monoclonal antibodies and nearly extra recombinant proteins. Other cell forms comprise numerous mouse myelomas, for example, NSO-murine myeloma cells, baby hamster kidney cell for the manufacture of cattle mouth and foot illness vaccine, monkey kidney cell used for polio vaccine and human cell line, for example, human embryonic kidney cells. Currently, Other investors have designated the two diverse hemagglutinins of H5N1 species based on the deactivating epitopes and reactivity through diverse neutralizing antibodies. The hemagglutinins of vaccines are separately articulated on the baculovirus packet (bivalent-BacHA) through its innate antigenic arrangement [Citation111]. CRISPR has empowered more exact and cost-effective bioengineering of mammalian cells. It remains a generally manual and time-consuming prepare. In addition, hereditary strategies are the fair one portion of the vital toolkit for the successful improvement of unused therapeutics. By applying its robotized foundry procedures to mammalian cell building – from DNA blend to systems-level investigation, quickening timelines and diminishing costs of the improvement cycle to create pharmaceutical’s like protein- or enzyme-based treatments [Citation112]. Elective bioengineering grafting tools that empower the exact and conditional tweak of splicing movement to change protein grouping, differing qualities, and eventually cellular behavior [Citation113].
7.2. Other pharmaceuticals important natural active compounds
The countless diversity of biomolecules formed by the filamentous fungus validates the manipulation of these microorganisms. In specific, the isolation with the recovery of taxol forming endophytic fungus is a novel and viable method for the yield of antineoplastic treatment. The expansion and usage of taxol forming fungus have made noteworthy development worldwide [Citation2]. Taxol is the most widely used antitumor agent that was industrialized in the past decades. It has been globally used for efficient cancer treatment. Recently, Taxol has been produced by Fusarium oxysporum, and Alternaria alternata MF5 (isolated from the bark of female Taxus yew) Aspergillus niger (from Taxus cuspidate), grown in different media and found to produce taxol through fermentation bioengineering for biopharmaceuticals [Citation2,Citation10,Citation114].
β-D-galactosidase is the biocatalyst accountable for the biocatalysts of lactose toward glucose along with galactose. The comprehensive marketplace for lactase has been growing suggestively because of its position in lactose prejudice action. Lactase is supplied in capsules or tablet to be applied as a food complements for people prejudiced to lactose earlier consumption of dairy products or milk. Another biotic drug of fungal importance is the asparaginase catalyst. This catalyst is applied for the action of designated forms of hematopoietic illnesses, for example, non-hodgkin lymphoma and acute lymphoblastic leukemia. Though, the drug, which is attained from Erwinia chrysanthemi and E. coli (ELSPARTM), causes simple immunological responses. Hence, the fungi could deliver an alternate to the prokaryotic enzymes as an antitumoral mediator as it has shown constancy and optimal pH adjoining physiological environments [Citation2]. However, the usage of filamentous fungus for the manufacture of compounds of attention is still a thought-provoking approach.
In the plant’s framework, the bioengineered retrosynthetic pathway plan deconstructs a plant common items one step at a time and utilizes reaction/enzyme sets from databases such as MetaCyc to propose biosynthetic courses to the target. RetroPath takes beginning compounds, a target, and response rules to create potential pathways and was tentatively approved in the plan of biosynthetic courses to pinocembrin, a flavonoid four enzymatic step from E. coli central digestion system [Citation115].
8. Application of bioengineered tools for pharmaceuticals productions
Bioengineered tools are confronting, thoughtful and solving extremely complex complications in biomedicine, bringing organized the tools accessible in the arenas of experimental science, life science and bioengineering in all their planes. Many investigators are used bioengineering tools for better recognize the proteins, tissue, cells and organs, or develop resolutions, for example, nanoparticles for specific drug delivery, nanotools for study biological organizations, molecular actuators that can be switched on and off with light, and in vitro organs ‘on-a-chip’ for disease models. Some of major important applications of the bioengineered tools are as follows:
8.1. In pharmacology and pharmaceuticals
Artemisia annua is a consideration of investigators since 1980s being a significant basis of antimalarial artemisinin drug and nonstop exertions are proceeding to an upsurge the manufacture of metabolite inside the plants and in homologous and heterologous arrangements. The beneficial forms of biopharmaceuticals are chiefly included the recombinant protein treatment, antibody therapy, gene therapy and cell therapy. Biopharmaceuticals are gifted at disease treatment securely and efficiently by indicating biological action and perform definite functions by stand-in on the illness pathophysiology [Citation116]. Presently, biopharmaceuticals are widely used as beneficial agents, for example, vaccines, whole blood, antigens, immunosera, hormones, enzymes, cytokines, allergenics, gene remedies, cell therapies, monoclonal antibodies and bioproducts resulting from recombinant DNA, etc. [Citation116]. As biotherapeutic protein examination using LC–MS tool is a developing zone, this distinct focus matter in the presence of advances and deliberations of application of LC-MS for characterization along with quantification of therapeutic proteins [Citation117]. Phenylalanine ammonia lyase has been derived from Rhodotorula rubia yeast, 4-Coumarate CoA ligase from bacteria S. coelicolor enzyme acetyl CoA carboxylase in addition to the stilbene synthase by Arachis hypogea. Additional, improved formation of pharmaceutically imperative polyunsaturated eicosapentaenoic acid, arachidonic acid is detected through employing nine genes collective composed to analyze a large quantity of denaturation and condensation reactions [Citation3]. For autoimmune illnesses, the transgenic plant, tobacco and potato by encoding gene for GAD67 have been established. The investigation has revealed that nourishing of diabetic mice with these vaccines aided in overpowering the autoimmune outbreak and overdue the increase in blood sugar stages [Citation118]. Valuation of mRNA S100A9 expression is a capable tool for the analysis of S100A9 and osteosarcoma that may be a capable healing target in osteosarcoma [Citation119]. Access-restricted commercial libraries will likely proceed to be a vital apparatus in normal items investigate within the future. A few later surveys have talked about the common capabilities of different MS-based databases and computer program apparatuses for pharmaceutical’s production. MetFrag is an Internet device that performs in silico fragmentations, permitting looks of exploratory MS spectra against chemical databases such as KEGG, PubChem, ChemSpider, or custom structure databases [Citation120].
8.2. In the industrial biotechnology sector
Bioengineering of scientifically imperative waxes, oils and supplementary plant lipids could show an amplified proportion of progression because of important upgrading in the fundamental tools. These could be demonstrated via safflower manufacture (Carthamus tinctorius L.) by higher oleic acid wherein the seed’s oil is metabolically bioengineered. It has covered the system for better utilization of oleic acids as a feedstock in addition to lubricants and in modifier fluids with trade insinuation offering a milestone utilization of metabolic bioengineering of extremely pure oleochemicals in plant oils to be applied as feedstock [Citation121]. Lavender (Lavandula latifolia red) is an imperative constituent of oil industry, refined all over the biosphere with limonene as a trivial component? A noteworthy upsurge of limonene contented in emerging leaves of spike lavender has been detected when the limonene synthase genes recovered from Mentha spicata are overexpressed under the regulator of the CaMV 35S constitutive promoter as associated with the control leaves [Citation122]. Similar limonene, the stage of other terpenoids (menthol, linalool, sesquiterpene and geraniol phytosterol) is also augmented effectively. An additional example of focused variation in metabolism through industrial operation is the alteration of fatty-acid configuration with triglycerides [Citation123].
9. Perspective, future outlook and challenges of bioengineered tools
Present situation of bioengineering tools be subject on the speedy development in molecular biology techniques and the expansion of innovative approaches for metabolic and genomic outlining. Integration of bioengineering with recently rising biosciences, has displayed a special opportunity to overcome major challenges to pharmacology, natural biology and human well-being [Citation124]. To saddle the potential of advanced bioengineering and setup a maintainable establishment for green innovation, researchers and engineers ought to be familiar with the regulating questions of science and innovation. Entirety cell biosensors devices give a basic strategy for the location of overwhelming metals as compared with past biosensors [Citation125]. Bioengineered bioelectronic instruments have picked up universal consideration within the past decade. Inveterate enactment of C-fibres may compound infection pathology in rheumatoid joint pain through the antidromic discharge of pro-inflammatory neuropeptides [Citation126]. An effective bioengineered crossbreed translation promoter through the combination of RegCG with CMV, which, in steady cell lines, appears more noteworthy action than when both promoters are utilized independently at that point other promoters [Citation127]. Hereditary circuits bioengineering devices have allowed a more focus on results and biomimetic reactions to neurotic conditions. Hereditary bioengineering apparatuses built by manufactured scientists empower tight control of quality expression that permits the energetic control of translation to calculate an expression in stem cells. These hereditary devices have been utilized for coordinating stem cell separation to create wanted cell ancestries, to form organoids, and to build restorative cells to sense and react to the malady [Citation128]. CRISPR/Cas9 framework may be a capable bioengineering tool for thumping out qualities in cells. The RNA-guided CRISPR/Cas9 framework may be an effective device for genome altering in assorted life forms and cell sorts [Citation129].
Recent expansion in the quantum information arriving as a result of tremendous evidence from the genome, transcriptome and proteome of several crops together with many aromatic and medicinal plants still needs many efforts to sharpening the tools more user-friendly for bioengineering. Though, there are still many challenges for assimilating the heterologous path in frame’s cells for huge production resolution due to its limited categorized portions, module’s unsuitability, and cell acceptance toward product [Citation130]. The bioengineered tools are advanced sophisticated tools for pharmaceuticals and other industry where it can be attained more authentic evidence for better diversities in horticulture, postharvest value, improved nutritional value, improved resistant variations, and high-value pharmaceutical compounds. However, along with the effective and inexpensive product, bioengineered tools are formed a lot of ethical, environmental and socioeconomical problems. As in few soybeans, altered cases by Brazil’s nut gene displays an allergic response subsequently feeding by cattle’s and such remarks necessitate a stricter examination.
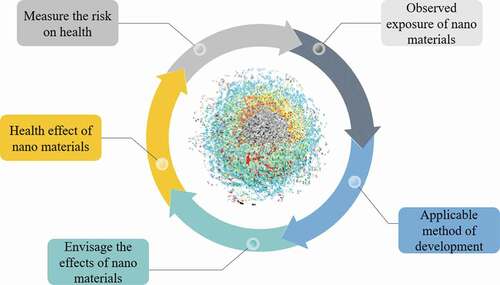
The perception of bioengineered tools is well recognized now for the formation of pharmaceuticals through the current agreement from food development authority on the newest investigation and expansion of the plant, animal and microbes-based production systems using system and synthetic biology with molecular biotechnological approaches [Citation1]. The formation of recombinant beneficial proteins is still expensive as a result of manufacturing difficulty of organism expression organizations, and the controlling burden related with managing these pharmaceutical’s drugs to the patients in a safer and effective manner. There is a prerequisite of additional effort on the protection problems of the genetically adapted organism and pharmaceutical’s production in apprehension to health and environment [Citation12]. It is important to protect intellectual property in all of these bioproducts through bioengineered tools because they are derived from unique genetic mutations. If we can obtain proof of application, the bioengineered tool’s and process could make living organism once again a major global source of pharmaceuticals, a position they have occupied throughout the history of [Citation14]. The major nano bioengineered tools challenges are yet to prevail over nano material’s measurement for the risk on health, nano material’s effects, observed exposure of nanomaterials, application methods, ecological hazards, manufacture cost and user-friendliness to the un-reachable at far-off zones ().
On the other hand, no research inventory analysis has been conducted on the life cycle assessment (LCA) of pharmaceutical’s bioproducts produced by bioengineered tools that these metrics could fail to the relevant decision-making factors that could be addressed more specifically by a critical comprehensive LCA. Deficiency of inventory information for many bioengineered tools and techniques poses an important barrier to more wide-ranging application of LCA for pharmaceuticals. A cradle-to-gate LCA of bioengineered tools dependent pharmaceuticals will be helpful for present the applied approach to the concept the inventories using research, optimization, production and literature data (). Apart from this, the growing data manipulated genes linked to metabolic paths, and through polished biotechnology in molecular level along with bioengineering could be a revolution the pharmaceuticals changing aspects of the entire biosphere. Novel developing computational biology simplifies the genetic bioengineering developments by dropping the period spent in manual examination. Amendment in gene expression, it could be found the transcriptional and/or posttranscriptional level, touch the complete metabolic outlining of plants. Bioengineered tools are backbone tools to fulfill the prerequisite of pharmaceutically demand of growing inhabitants.
10. Conclusions
In the present review, the bioengineered tools are widely depending on the speedy development in system biology, synthetic biology along with biotechnological molecular biology tools and techniques for the development of new approaches for pharmaceuticals outlining. It concluded that new tools and technological developments are endlessly made to expand the discovery, rational variation, production and purification of important biopharmaceuticals. The preliminary LCA models can also be a valuable tool to screen potential bioengineered alternatives. For the production of pharmaceutically and industrially valuable compounds, bioengineering is a magical tool which can fill the gap of global pharmaceutical’s bioproducts demands.
Highlights
Given new insights on the development of bioengineered tools for the pharmaceutical’s bioproducts;
Integrations of transcriptome, proteome, metabolome and nano bioengineering tools are useful;
Critically described the challenges and future possibilities of the bioengineering tools.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Khatodia S, Khurana SMP. Genetic engineering for plant transgenesis. In: omics Technologies and Bio-Engineering. Elsevier; 2018. p. 71–86. doi:https://doi.org/10.1016/b978-0-12-815870-8.00005-x.
- Jozala AF, Geraldes DC, Tundisi LL, et al. Biopharmaceuticals from microorganisms: from production to purification. Braz J Microbiol. 2016;47:51–63.
- Sangwan NS, Jadaun JS, Tripathi S, et al. Plant Metabolic Engineering. In: Omics Technologies and Bio-Engineering. Elsevier; 2018. p. 143–175. doi:https://doi.org/10.1016/b978-0-12-815870-8.00009-7.
- Kalia A. Chapter 12 - Nanotechnology in Bioengineering: transmogrifying Plant Biotechnology. In: Barh D, Azevedo V, editors. Omics Technologies and Bio-Engineering. Academic Press; 2018. p. 211–229. doi:https://doi.org/10.1016/B978-0-12-815870-8.00012-7.
- Wang G, Huang M, Nielsen J. Exploring the potential of Saccharomyces cerevisiae for biopharmaceutical protein production. Curr Opin Biotech. 2017 Dec;48:77–84..
- Patil RH, Patil MP, Maheshwari VL. Bioactive secondary metabolites from endophytic fungi. In: Studies in natural products chemistry. Elsevier; 2016. p. 189–205. doi:https://doi.org/10.1016/b978-0-444-63601-0.00005-3.
- Xu N, Zhu J, Q Z, et al. Identification and characterization of novel promoters for recombinant protein production in yeast Pichia pastoris. Yeast. Wiley. 2018 8;35(5):379–385.
- Qiao W, Ling F, Yu L, et al. Enhancing taxol production in a novel endophytic fungus, Aspergillus aculeatinus Tax-6, isolated from Taxus chinensis var. mairei. Fungal Biol. 2017;121(12):1037–1044.
- Kumaran RS, Kim HJ, Hur B-K. Taxol promising fungal endophyte, Pestalotiopsis species isolated from Taxus cuspidata. J Biosci Bioeng. 2010;110(5):541–546.
- Yang N, Pan X, Chen G, et al. Fermentation engineering for enhanced paclitaxel production by taxus media endophytic fungus MF-5 (Alternaria sp.). J Biobased Mater Bio. 2018;12(6):545–550.
- Sudhakar N, Shanmugam H, Kumaran S, et al. Omics approaches in fungal biotechnology. In Omics Technologies and Bio-Engineering. Elsevier; 2018. p. 53–70. doi:https://doi.org/10.1016/b978-0-12-815870-8.00004-8.
- Tzani I, Monger C, Kelly P, et al. Understanding biopharmaceutical production at single nucleotide resolution using ribosome footprint profiling. Curr Opin Biotech. 2018;53:182–190.
- Kumar S, Dangi AK, Shukla P, et al. Thermozymes: adaptive strategies and tools for their biotechnological applications. Bioresour Technol. 2019;278:372–382.
- Brown DP, Rogers DT, Gunjan SK, et al. Target-directed discovery and production of pharmaceuticals in transgenic mutant plant cells. J Biotechnol. 2016;238:9–14.
- Breitling R, Takano E. Synthetic biology advances for pharmaceutical production. Curr Opin Biotech. 2015;35:46–51.
- Li H, Shen CR, Huang CH, et al. CRISPR-Cas9 for the genome engineering of cyanobacteria and succinate production. Metab Eng. 2016;38:293–302.
- Wang J, Kong S, Chen F, et al. A bioelectronic taste sensor based on bioengineered Escherichia coli cells combined with ITO-constructed electrochemical sensors. Anal Chim Acta. 2019. doi:https://doi.org/10.1016/j.aca.2019.06.023.
- Amitai G, Sorek R. PanDaTox. Bioengineered. 2012;3(4):218–221.
- Nakamae K, Nishimura Y, Takenaga M, et al. Establishment of expanded and streamlined pipeline of PITCh knock-in - a web-based design tool for MMEJ-mediated gene knock-in, PITCh designer, and the variations of PITCh, PITCh-TG and PITCh-KIKO. Bioengineered. 2017;8(3):302–308.
- Ventura M, Turroni F, van Sinderen D. Probiogenomics as a tool to obtain genetic insights into adaptation of probiotic bacteria to the human gut. Bioengineered. 2012;3(2):73–79.
- Dziadek B, Brzostek A. Recombinant ROP2, ROP4, GRA4 and SAG1 antigen-cocktails as possible tools for immunoprophylaxis of toxoplasmosis. Bioengineered. 2012;3(6):358–364.
- Kick L, Kirchner M, Schneider S. CRISPR-Cas9: from a bacterial immune system to genome-edited human cells in clinical trials. Bioengineered. 2017;8(3):280–286.
- Khatodia S, Bhatotia K, Tuteja N. Development of CRISPR/Cas9 mediated virus resistance in agriculturally important crops. Bioengineered. 2017;8(3):274–279.
- Nakade S, Yamamoto T, Sakuma T. Cas9, Cpf1 and C2c1/2/3—What’s next? Bioengineered. 2017;8(3):265–273.
- Li Y, Li S, Li Y, et al. Generation of a novel HEK293 luciferase reporter cell line by CRISPR/Cas9-mediated site-specific integration in the genome to explore the transcriptional regulation of the PGRN gene. Bioengineered. 2019;10(1):98–107.
- Perez RF, Nyman RKM, Johnson AAT, et al. CRISPR-Cas systems: ushering in the new genome editing era. Bioengineered. 2018;9(1):214–221.
- Al-Omari S, Mubarak AA, Al-Noaimi M, et al. Multielemental analysis of pharmaceuticals derived from plant seeds by energy dispersive X-ray fluorescence spectrometry. Instrum Sci Technol. 2015;44(1):98–113.
- Schofield DA, Bull CT, Rubio I, et al. “Light-tagged” bacteriophage as a diagnostic tool for the detection of phytopathogens. Bioengineered. 2013;4(1):50–54.
- Sarsaiya S, Jia Q, Fan X, et al. First report of leaf black circular spots on Dendrobium nobile caused by Trichoderma longibrachiatum in Guizhou Province, China. Plant Dis. 2019. doi:https://doi.org/10.1094/pdis-03-19-0672-pdn.
- Narnoliya LK, Kaushal G, Singh SP, et al. De novo transcriptome analysis of rose-scented geranium provides insights into the metabolic specificity of terpene and tartaric acid biosynthesis. BMC Genomics. 2017;18(1). DOI:https://doi.org/10.1186/s12864-016-3437-0
- Mashima J, Kodama Y, Fujisawa T, et al. DNA Data Bank of Japan. Nucleic Acids Res. 2017;45:D25–D31.
- Baker W, van Den Broek A, Camon E, et al. The EMBL nucleotide sequence database. Nucleic Acids Res. 2000;28(1):19–23.
- Buchmann JP, Holmes EC. Entrezpy: A Python library to dynamically interact with the NCBI Entrez databases. Bioinformatics. 2019;11:btz385.
- Mukherjee S, Stamatis D, Bertsch J, et al. Genomes online database (GOLD) v.6: data updates and feature enhancements. Nucleic Acids Res. 2017;45:D446–D456.
- Clough E, Barrett T. The gene expression omnibus database. Methods Mol Biol. 2016;1418:93–110.
- Parkinson H, Kapushesky M, Shojatalab M, et al. ArrayExpress—a public database of microarray experiments and gene expression profiles. Nucleic Acids Res. 2007;35:D747–D750.
- Gasteiger E, Gattiker A, Hoogland C, et al. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31(13):3784–3788.
- Mewes HW, Frishman D, Güldener U, et al. MIPS: a database for genomes and protein sequences. Nucleic Acids Res. 2002;30(1):31–34.
- von Mering C, Huynen M, Jaeggi D, et al. STRING: A database of predicted functional associations between proteins. Nucleic Acids Res. 2003;31(1):258–261.
- Bairoch A. The ENZYME database in 2000. Nucleic Acids Res. 2000;28(1):304–305.
- Schomburg I, Chang A, Schomburg D. BRENDA, enzyme data and metabolic information. Nucleic Acids Res. 2002;30(1):47–49.
- Karp PD, Billington R, Holland TA, et al. Computational metabolomics operations at BioCyc.org. Metabolites. 2015;5(2):291–310.
- Gu Y, Yang D, Zou J, et al. Systematic interpretation of comutated genes in large-scale cancer mutation profiles. Mol Cancer Ther. 2010;9(8):2186–2195.
- Kanehisaa M, KEGG: GS. Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30.
- Caspi R, Billington R, Fulcher CA, et al. The MetaCyc database of metabolic pathways and enzymes. Nucleic Acids Res. 2018;46:D633–D639.
- Blanchard JL, Bulmore DL, Farmer AD, et al. PathDB: A second generation metabolic database; 2019. Available from: https://pdfs.semanticscholar.org/ecec/4ad703875624328c49f8debca05d5291b860.pdf
- Gangrade D, Sawant G, Mehta A. Re-thinking drug discovery: in silico method. J Chem Pharmaceutical Res. 2016;8(8):1092–1099.
- Klamt S, Stelling J, Ginkel M, et al. FluxAnalyzer: exploring structure, pathways, and flux distributions in metabolic networks on interactive flux maps. Bioinformatics. 2003;19(2):261–269.
- Thiele I, Palsson BØ. A protocol for generating a high-quality genome-scale metabolic reconstruction. Nat Protoc. 2010;5(1):93–121.
- Kolisnychenko V. Engineering a reduced escherichia coli genome. Genome Res. 2002;12(4):640–647.
- Rastogi S, Meena S, Bhattacharya A, et al. De novo sequencing and comparative analysis of holy and sweet basil transcriptomes. BMC Genomics. 2014;15(1):588–607.
- Brandan CP, Basombrío MÁ. Genetically attenuated Trypanosoma cruzi parasites as a potential vaccination tool. Bioengineered. 2012;3(4):242–246.
- Tang Q, Ma X, Mo C, et al. An efficient approach to finding Siraitia grosvenorii triterpene biosynthetic genes by RNA-seq and digital gene expression analysis. BMC Genomics. 2011;12(1):343.
- Robertson G, Schein J, Chiu R, et al. De novo assembly and analysis of RNA-seq data. Nat Methods. 2010;7:909–912.
- Weckwerth W. Integration of metabolomics and proteomics in molecular plant physiology coping with the complexity by data-dimensionality reduction. Physiol Plant. 2008;132:176–189.
- Yamaguchi H, Miyazaki M. Microfluidic chips with multi-junctions: an advanced tool in recovering proteins from inclusion bodies. Bioengineered. 2015;6(1):1–4.
- Huchelmann A, Boutry M, Hachez C. Plant glandular trichomes: natural cell factories of high biotechnological interest. Plant Physiol. 2017;175(1):6–22.
- Yoon JM, Zhao L, Shanks JV. Metabolic engineering with plants for a sustainable biobased economy. Annu Rev Chem Biomol Eng. 2013;4:211–237.
- Hou BK, Kim JS, Jun JH, et al. BioSilico: an integrated metabolic database system. Bioinformatics. 2004;20(17):3270–3272.
- Radrich K, Tsuruoka Y, Dobson P, et al. Integration of metabolic databases for the reconstruction of genome-scale metabolic networks. BMC Syst Biol. 2010;4:114.
- Granata I, Troiano E, Sangiovanni M, et al. Integration of transcriptomic data in a genome-scale metabolic model to investigate the link between obesity and breast cancer. BMC Bioinformatics. 2019;20(4):162.
- Jia Z. Barnes Mcogena: co-expressed gene-set enrichment analysis. R package version 1.18.0; 2019. Available from: https://github.com/zhilongjia/cogena
- Lagunin A, Ivanov S, Rudik A, et al. DIGEP-Pred: web service for in silico prediction of drug-induced gene expression profiles based on structural formula. Bioinformatics. 2013;29(16):2062–2063.
- Laenen G, Ardeshirdavani A, Moreau Y, et al. Galahad: a web server for drug effect analysis from gene expression. Nucleic Acids Res. 2015;43:W208- W212.
- Napolitano F, Carrella D, Mandriani B, et al. Gene2drug: a computational tool for pathway-based rational drug repositioning. Bioinformatics. 2018;34(9):1498–1505.
- Brown AS, Kong SW, Kohane IS, et al. ksRepo: A generalized platform for computational drug repositioning. BMC Bioinformatics. 2016;17:78.
- Carrella D, Napolitano F, Rispoli R, et al. Mantra 2.0: an online collaborative resource for drug mode of action and repurposing by network analysis. Bioinformatics. 2014;30(12):1787–1788.
- Luo H, Li M, Wang S, et al. Computational drug repositioning using low-rank matrix approximation and randomized algorithms. Bioinformatics. 2018;34(11):1904–1912.
- Navarro C, Martínez V, Blanco A, et al. ProphTools: general prioritization tools for heterogeneous biological networks. Gigascience. 2017;6(12):1–8.
- Backman TW, Cao Y, Girke T. ChemMine tools: an online service for analyzing and clustering small molecules. Nucleic Acids Res. 2011;39:W486–91.
- Ravikumar B, Alam Z, Peddinti G1, et al. C-SPADE: a web-tool for interactive analysis and visualization of drug screening experiments through compound-specific bioactivity dendrograms. Nucleic Acids Res. 2017;45(W1):W495–W500.
- Cobanoglu MC1, Oltvai ZN1, Taylor DL1, et al. BalestraWeb: efficient online evaluation of drug-target interactions. Bioinformatics. 2015;31(1):131–133.
- Yamanishi Y1, Kotera M2, Moriya Y3, et al. DINIES: drug-target interaction network inference engine based on supervised analysis. Nucleic Acids Res. 2014;42:W39–45.
- Alaimo S, Bonnici V, Cancemi D, et al. DT-Web: a web-based application for drug-target interaction and drug combination prediction through domain-tuned network-based inference. BMC Syst Biol. 2015;9(3):S4.
- Awale M, Reymond JL. The polypharmacology browser: a web-based multi-fingerprint target prediction tool using ChEMBL bioactivity data. J Cheminf [Internet]. Springer Science and Business Media LLC. 2017 21;9(1). doi:https://doi.org/10.1186/s13321-017-0199-x.
- Cañada A, Capella-Gutierrez S, Rabal O, et al. LimTox: a web tool for applied text mining of adverse event and toxicity associations of compounds, drugs and genes. Nucleic Acids Res. 2017;45(W1):W484–W489.
- King ZA, Lu J, Dräger A, et al. BiGG Models: A platform for integrating, standardizing and sharing genome-scale models. Nucleic Acids Res. 2016;44:D515–D522.
- Lee SY, Lee DY, Hong SH, et al. MetaFluxNet, a program package for metabolic pathway construction and analysis, and its use in large-scale metabolic flux analysis of Escherichia coli. Genome Inform. 2003;14:23–33.
- Adrio J-L, Demain AL. Recombinant organisms for production of industrial products. Bioengineered Bugs. 2010;1(2):116–131.
- Dwane S, Kiely PA. Tools used to study how protein complexes are assembled in signaling cascades. Bioengineered Bugs. 2011;2(5):247–259.
- Santos-Merino M, Singh AK, Ducat DC. New applications of synthetic biology tools for cyanobacterial metabolic engineering. Front Bioeng Biotechnol. 2019;7:33.
- Jadaun JS, Sangwan NS, Narnoliya LK, et al. Over-expression of DXS gene enhances terpenoidal secondary metabolite accumulation in rose-scented geranium and Withania somnifera: active involvement of plastid isoprenogenic pathway in their biosynthesis. Physiol Plant. 2017a;159(4):381–400.
- Bekker V, Dodd A, Brady D, et al. Tools for metabolic engineering in Streptomyces. Bioengineered. 2014;5(5):293–299.
- Charlton Hume HK, Vidigal J, Carrondo MJT, et al. Synthetic biology for bioengineering virus-like particle vaccines. Biotechnol Bioeng. 2018;116(4):919–935.
- Sanzari I. Leone A and Ambrosone A. Nanotechnology in plant science: to make a long story short. Front Bioeng Biotechnol. 2019;7:1–12.
- Hemelaar SR, van der Laan KJ, Hinterding SR, et al. Generally applicable transformation protocols for fluorescent nanodiamond internalization into cells. Sci Rep. 2017;7:5862.
- Cunningham FJ, Goh NS, Demirer GS, et al. Nanoparticle-mediated delivery towards advancing plant genetic engineering. Trends Biotechnol. 2018;36(9):882–897.
- Martin-Ortigosa S, Wang K. Proteolistics: a biolistic method for intracellular delivery of proteins. Transgenic Res. 2014;23(5):743–756.
- Senol Cali D, Kim JS, Ghose S, et al. Nanopore sequencing technology and tools for genome assembly: computational analysis of the current state, bottlenecks and future directions. Brief Bioinform. 2018. DOI:https://doi.org/10.1093/bib/bby017
- Bowden R, Davies RW, Heger A, et al. Sequencing of human genomes with nanopore technology. Nat Commun. 2019;10(1). doi:https://doi.org/10.1038/s41467-019-09637-5
- Sanderson ND, Street TL, Foster D, et al. Real-time analysis of nanopore-based metagenomic sequencing from infected orthopaedic devices. BMC Genomics. 2018;19(1). doi:https://doi.org/10.1186/s12864-018-5094-y
- Sharma S, Jaiswal S, Duffy B, et al. Nanostructured materials for food applications: spectroscopy, microscopy and physical properties. Bioengineering. 2019 19;6(1):26.
- Martinelli C, Pucci C, Ciofani G. Nanostructured carriers as innovative tools for cancer diagnosis and therapy. APL Bioeng. 2019;3(1):011502.
- Ferenz K, Zhang C. Nano-bio-imaging and therapeutic nanoparticles. J Nanosci Nanomed. 2018;2(1):19–20.
- Siddiqui MH, Al-Whaibi MH, Firoz M, et al. Role of nanoparticles in plants. Nanotechnol Plant Sci. 2015;19–35. DOI:https://doi.org/10.1007/978-3-319-14502-0_2
- Yah CS, Simate GS. Nanoparticles as potential new generation broad spectrum antimicrobial agents. DARU J Pharm Sci. 2015;23:43.
- Lim EK, Kim T, Paik S, et al. Nanomaterials for theranostics: recent advances and future challenges. Chem Rev. 2015;115:327394.
- Ozkan B, Budama-Kilinc Y, Cakir-Koc R, et al. Application of an immunoglobulin Y-alkaline phosphatase bioconjugate as a diagnostic tool for influenza A virus. Bioengineered. 2019;10(1):33–42.
- Geiss GK, Bumgarner RE, Birditt B, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008;26(3):317325.
- Chen K, Han H, Luo Z, et al. A practicable detection system for genetically modified rice by SERS-barcoded nanosensors. Biosens Bioelectron. 2012;34:118–124.
- Shikha S, Salafi T, Cheng J, et al. Versatile design and synthesis of nano-barcodes. Chem Soc Rev. 2017;46(22):7054–7093.
- Lokugamage MP, Sago CD, Dahlman JE. Testing thousands of nanoparticles in vivo using DNA barcodes. Curr Opin Biomed Eng. 2018;7:1–8.
- Chaudhary B, Singh N, Pandey DK. Bioengineering of crop plants for improved tetrahydrofolate production. Bioengineered. 2018;9(1):152–158.
- Lalor F, Fitzpatrick J, Sage C, et al. Sustainability in the biopharmaceutical industry: seeking a holistic perspective. Biotechnol Adv. 2019;37(5):698–707.
- Baumgarten T, Ytterberg AJ, Zubarev RA, et al. Optimizing recombinant protein production in the Escherichia coli periplasm alleviates stress. Appl Environ Microbiol. 2018;84(12):e00270–18.
- Routledge SJ, Mikaliunaite L, Patel A, et al. The synthesis of recombinant membrane proteins in yeast for structural studies. Methods. 2016;95:26–37.
- Çelik E, Çalık P. Production of recombinant proteins by yeast cells. Biotechnol Adv. 2012;30(5):1108–1118.
- Love KR, Dalvie NC, Love JC. The yeast stands alone: the future of protein biologic production. Curr Opin Biotech. 2018;53:50–58.
- Vieira Gomes A, Souza Carmo T, Silva Carvalho L, et al. Comparison of yeasts as hosts for recombinant protein production. Microorganisms. 2018;6(2):38.
- Gidijala L, Uthoff S, van Kampen SJ, et al. Presence of protein production enhancers results in significantly higher methanol-induced protein production in Pichia pastoris. Microb Cell Fact. 2018;17(1). DOI:https://doi.org/10.1186/s12934-018-0961-4
- Prabakaran M, Kwang J. Recombinant baculovirus displayed vaccine. Bioengineered. 2014;5(1):45–48.
- Black JB, Perez-Pinera P, Gersbach CA. Mammalian synthetic biology: engineering biological systems. Annu Rev Biomed Eng. 2017;19(1):249–277.
- Mathur M, Xiang JS, Smolke CD. Mammalian synthetic biology for studying the cell. J Cell Biol. 2016;216(1):73–82.
- Sarsaiya S, Shi J, Chen J. A comprehensive review on fungal endophytes and its dynamics on Orchidaceae plants: current research, challenges, and future possibilities. Bioengineered. 2019;10(1):316–334.
- Cravens A, Payne J, Smolke CD. Synthetic biology strategies for microbial biosynthesis of plant natural products. Nat Commun. 2019;10(1). DOI:https://doi.org/10.1038/s41467-019-09848-w
- Chen YC, Yeh MK. Introductory Chapter: biopharmaceuticals. Biopharmaceuticals. 2018;19. doi:https://doi.org/10.5772/intechopen.79194.
- Zhang YJ, An HJ. Technologies and strategies for bioanalysis of biopharmaceuticals. Bioanalysis. 2017;9(18):1343–1347.
- Munshi A, Sharma V. Omics and edible vaccines. In: Omics Technologies and Bio-Engineering. Elsevier; 2018. p. 129–141. doi:https://doi.org/10.1016/b978-0-12-815870-8.00008-5.
- Liu Y, Luo G, He D. Clinical importance of S100A9 in osteosarcoma development and as a diagnostic marker and therapeutic target. Bioengineered. 2019;10(1):133–141.
- Johnson SR, Lange BM. Open-access metabolomics databases for natural product research: present capabilities and future potential. Front Bioeng Biotech. 2015;3. doi:https://doi.org/10.3389/fbioe.2015.00022.
- Škorić D, Jocić S, Sakač Z, et al. Genetic possibilities for altering sunflower oil quality to obtain novel oils. Can J Physiol Pharmacol. 2008;86:215–221.
- Muñoz-Bertomeu J, Ros R, Arrillaga I, et al. Expression of spearmint limonene synthase in transgenic spike lavender results in an altered monoterpene composition in developing leaves. Metab Eng. 2008;10:166–177.
- Voelker T, Worrell A, Anderson L, et al. Fatty-acid biosynthesis redirected to medium chains in transgenic oilseed plants. Science. 1992;257:72–74.
- Jain A, Sarsaiya S, Wu Q, et al. New insights and rethinking of cinnabar for chemical and its pharmacological dynamics. Bioengineered. 2019;10(1):353–364.
- Guo M, Du R, Xie Z, et al. Using the promoters of MerR family proteins as “rheostats” to engineer whole-cell heavy metal biosensors with adjustable sensitivity. J Biol Eng. 2019;13(1). DOI:https://doi.org/10.1186/s13036-019-0202-3
- Seicol BJ, Bejarano S, Behnke N, et al. Neuromodulation of metabolic functions: from pharmaceuticals to bioelectronics to biocircuits. J Biol Eng. 2019;13(1). DOI:https://doi.org/10.1186/s13036-019-0194-z
- Zúñiga RA, Gutiérrez-González M, Collazo N, et al. Development of a new promoter to avoid the silencing of genes in the production of recombinant antibodies in chinese hamster ovary cells. J Biol Eng. 2019;13(1). DOI:https://doi.org/10.1186/s13036-019-0187-y
- Healy CP, Deans TL. Genetic circuits to engineer tissues with alternative functions. J Biol Eng. 2019;13(1). DOI:https://doi.org/10.1186/s13036-019-0170-7
- Wang B, Wang Z, Wang D, et al. krCRISPR: an easy and efficient strategy for generating conditional knockout of essential genes in cells. J Biol Eng. 2019;13(1). DOI:https://doi.org/10.1186/s13036-019-0150-y
- Li J, Meng H, Wang Y. Synbiological systems for complex natural products biosynthesis. Synth Syst Biotechnol. 2016;1(4):221–229.