ABSTRACT
In preeclampsia (PE), preexistent maternal endothelial dysfunction leads to impaired placentation and vascular maladaptation. Long noncoding RNAs (lncRNAs) have been shown to participate in the placentation process. LncRNA fms-related tyrosine kinase 1 pseudogene 1 (FLT1P1) was previously reported to be upregulated in PE. In this study, we verified the effect of FLT1P1 and its cognate gene FLT1 on trophoblast cell proliferation and angiogenesis by using Cell Counting Kit-8 (CCK-8) assay, tube formation assay, and western blot analysis. Their binding to RNA binding protein dyskeratosis congenita 1 (DKC1) was validated by conducting RNA immunoprecipitation (RIP) and RNA pulldown assays. In this study, knockdown of FLT1P1 or FLT1 was found to promote cell proliferation and angiogenesis in trophoblasts. In addition, FLT1P1 recruited DKC1 to stabilize FLT1. Importantly, silencing FLT1P1 or DKC1 decreased the stability of FLT1. Rescue assays revealed that FLT1 overexpression reversed the effect of silenced FLT1P1. Overall, FLT1P1 cooperates with DKC1 to restrain trophoblast cell proliferation and angiogenesis by targeting FLT1.
KEYWORDS:
Introduction
Preeclampsia (PE), with a global prevalence of estimated 2% to 8% of all pregnancies, is one of the causes of maternal and fetal mortality [Citation1]. It is related to abnormal placentation and maladaptation of the maternal cardiovascular system, in which vascular resistance is abnormally increased, resulting in maternal hypertension [Citation2]. Importantly, the disease is linked to maternal long-term cardiovascular disorders and systemic inflammation [Citation3]. PE is a consequence of various pathophysiological processes including altered trophoblast proliferation, reduced invasion of extravillous trophoblasts, impaired differentiation of trophoblastic cells, and dysregulated immunoregulation [Citation4,Citation5]. However, its pathogenesis and associated molecular mechanisms are still not completely understood.
Long non-coding RNAs (lncRNAs) are defined as transcribed RNA molecules ranging from 200 to 100,000 nucleotides that do not code for any protein [Citation6]. It has been recognized that lncRNAs play different roles in many important biological processes, including regulation of transcriptional and posttranscriptional processes, epigenetic control, differentiation and development, cell cycle control, apoptosis, and metabolic processes [Citation7], participating in the pathogenesis and development of various diseases, including PE [Citation8,Citation9]. Recently, lncRNAs were identified to be associated with the pathogenesis of PE. For example, downregulation of lncRNA maternally expressed gene 3 (MEG3) promotes the apoptosis and suppresses the migration of trophoblast cells [Citation10]. LncRNA metastasis associated lung adenocarcinoma transcript-1 (MALAT-1) is expressed at a low level in PE and regulates JEG-3 trophoblast cell migration and invasion [Citation11]. As a subtype of lncRNA, pseudogene shares high sequence homology with its cognate gene, which has the capacity to code protein [Citation12]. It was also found that pseudogene could regulate its cognate gene. As reported, the pseudogene phosphatase and tensin homolog pseudogene 1 (PTENP1) modulate the level of its matched protein-coding gene phosphatase and tensin homolog (PTEN) [Citation13]. Since then, numerous pseudogenes have been validated to exert critical functions in diverse pathophysiological and physiological processes [Citation14,Citation15]. There are few reports concerning the roles of pseudogenes in PE. The pseudogene urate (hydroxyiso-) hydrolase, pseudogene (URAHP), promotes proliferation and regulates the pathogenesis of PE [Citation16]. Dysfunction of pseudogene phosphoglycerate kinase 1, pseudogene 2 (PGK1P2) is involved in PE by acting as a competing endogenous RNA of phosphoglycerate kinase 1 [Citation17]. The structural similarity to lncRNA makes the pseudogene modulate its target gene expression by recruiting RNA binding proteins (RBPs) at the transcriptional level [Citation18,Citation19]. Dyskeratosis congenita 1 (DKC1) is a nucleolar, RBP, which is highly conserved in eukaryotes [Citation20]. This protein is a key component of the telomerase complex and an essential structural subunit of the telomerase ribonucleoprotein and it has the ability to activate telomerase ribonucleoprotein activity, resulting in telomere shortening [Citation21]. Previous studies have shown that DKC1 as a RBP is negatively regulated by MEG3 that has been frequently reported to promote trophoblast migration and decrease apoptosis in PE [Citation22–24]. We found a novel lncRNA, FLT1P1 (fms-related tyrosine kinase 1 pseudogene 1), as the pseudogene of VEGFR1 (vascular endothelial growth factor receptor-1), which is also known as FLT1 (fms-related tyrosine kinase 1) [Citation25]. In addition, FLT1P1 was reported to be overexpressed in preeclamptic placentas [Citation26], but its function and molecular mechanism remain unclear in PE.
In this study, we focused on identifying the role of the pseudogene FLT1P1 in trophoblast cell proliferation and angiogenesis and mechanistically analyzing how FLT1P1 exerts its function in PE. We hypothesized that FLT1P1 may exert its function in PE by interacting with the RBP DKC1 to stabilize its cognate gene FLT1. Our study may provide a novel regulatory mechanism for exploration of the pathogenesis of PE.
Materials and methods
Samples
Normal placentas (n = 10) were obtained from full-term births after the cesarean section. Age-matched placentas were obtained from women with severe PE (n = 10) after cesarean section. All placentas involved in this study were collected by procedures of planned cesarean section without the aid of artificial labor. Placental tissues were obtained by a certified doctor by making a vertical incision across a normal area at the center, involving fetal and maternal placental surfaces. Tissues having calcified deposits or clots were excluded. The experiments were approved by the Ethics Committee of the Affiliated Huaian No. 1 People’s Hospital of Nanjing Medical University. All volunteers participating in this study signed written informed consent.
Isolation and culture of human primary trophoblast cells (PTBs)
Term placentas were collected from uncomplicated pregnancies after cesarean delivery. Isolation and culture of PTBs was done according to conventional methods. Specifically, we used a protocol based on the classic trypsin digestion and Percoll gradient centrifugation method, as previously described [Citation27]. Briefly, the placental tissues were washed, sheared, weighed, and digested in a solution containing 0.125% trypsin and 0.03% DNase (Sigma, USA). The supernatant was collected, and the pellet was kept. This process was performed in order to discard the outer syncytium and to keep the underlying trophoblasts. The pellet was then purified with 5%–65% Percoll density gradients (Sigma), which allowed the collection of the trophoblasts. Finally, the trophoblasts were cultured in Dulbecco’s modified Eagle’s medium (Thermo Fisher Scientific, USA) containing 10% fetal bovine serum (FBS).
Cell culture
HTR-8/SVneo (HTR8) and BeWo cell lines were obtained from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). Cells were cultured in Roswell Park Memorial Institute (RPMI) 1640 (HyClone, USA) supplemented with 100 U/ml penicillin (HyClone), 10% heat-inactivated FBS, and 100 μg/ml streptomycin (Invitrogen, USA) in a humidified incubator at 37°C with 5% CO2.
Cell transfection
The short hairpin RNAs (shRNAs) against FLT1P1, FLT1 or DKC1 (sh-FLT1P1, sh-FLT1 or sh-DKC1) and the scrambled negative control (sh-NC) were designed and synthesized by GenePharma (Shanghai, China). The plasmid vector expressing full-length FLT1 was generated by GenePharma to overexpress FLT1 and termed as pcDNA3.1/FLT1 (FLT1), and the empty vector (vector) was used as a negative control. Cells were seeded in 24-well plates at 2 × 105 cells/well and transfected with 40 nM shRNA vector or 0.2 μg overexpression vector following the instructions of Lipofectamine 2000 (Invitrogen, USA) as described previously [Citation28], and cells were harvested at 48 h for further analysis.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
TRIzol (Invitrogen, USA) was used to isolate total RNA from trophoblast cells. Then, the first-strand cDNA was generated by ImProm-II Reverse Transcription System (Promega, USA). RT-qPCR was conducted by using SYBR Green qPCR assay (Takara, Dalian, China) and gene-specific primers. The relative gene expression was calculated by the 2−ΔΔCt method [Citation29], and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as the internal control. FLT1P1, forward: 5ʹ-AAGAACGCCGATTATGTGAG-3ʹ, reverse: 5ʹ-CAAGAGCCACCCATTTCAG-3ʹ; FLT1, forward: 5ʹ-CAAGATTGACTTGAGAGTAACCAG-3ʹ, reverse: 5ʹCTGGAATGGCAGAAACTGG-3ʹ; DKC1, forward: 5ʹ-GGTATAGTAGCCAAGATCAAGAG-3ʹ, reverse: 5ʹ-TTCTGACTTGCCTTTGGAC-3ʹ; GAPDH, forward: 5ʹ-TCAAGATCATCAGCAATGCC-3ʹ, reverse: 5ʹ-CGATACCAAAGTTGTCAT GGA-3ʹ.
Cell counting kit-8 (CCK-8) assay
The cell Counting Kit-8 (CCK-8, Dojindo Molecular Technologies, Kyushu, Japan) was used to measure the viability of stably transfected trophoblast cells. In brief, cells were seeded in 96-well plates at 4 × 103 cell/well. At 24 h, 48 h, and 72 h, each well was added with 10 μl CCK-8 solution for an additional 1 h of incubation at 37°C. Finally, a microplate reader (BioRad, CA, USA) was used to read the absorbance value at 450 nm [Citation30].
Tube formation assay
For the tube formation assay, as described previously [Citation31], 24-well plate was coated with growth factor reduced Matrigel (60 µl, Corning, NY, USA) for 1 h at 37°C. A total of 1 × 105 trophoblast cells in a medium containing 10% FBS were plated on top of presolidified Matrigel. Once seeded on Matrigel, capillary tubes and networks start to form. After 6 h of incubation, plates were examined with a microscope (Nikon, Japan), and images were taken. The number of branching points was quantified using ImageJ plug-in according to the protocol angiogenesis analyzer from Gilles Carpentier.
Western blot analysis
Western blot was performed as previously published [Citation32]. Total protein was extracted from cells using radioimmunoprecipitation assay lysis buffer (Life Technologies, USA) containing protease inhibitors (Sigma). After that, protein concentration was quantified with the Bicinchoninic Acid Assay (Beyotime, China). Before transferring into polyvinylidene difluoride membranes (Millipore, Billerica, MA, USA), the protein samples were separated with 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis. After blocking with 5% skim milk, the membranes were incubated with primary antibodies including vascular endothelial growth factor A (VEGFA; ab214424, 1:1000, Abcam, Cambridge, USA), fibroblast growth factor 2 (FGF2; ab208687, 1:1000), transforming growth factor beta (TGF-β; ab124894, 1:1000), FLT1 (ab32152, 1:1000), and GAPDH (ab181602, 1:10,000) overnight at 4°C. After washing, the membranes were further incubated with secondary antibodies (ab205718, 1:3000) for 1 h at room temperature. The blots were detected by using a chemiluminescence substrate (Pierce, USA). Immunoblot signals were quantified using Image Quant software (GE Healthcare).
RNA immunoprecipitation (RIP) assay
For the RIP assay, the Magna RIP TM RNA-Binding Protein Immunoprecipitation Kit (Millipore, Billerica, USA) was used following the manufacturer’s protocols [Citation28]. The trophoblast cells at 80–90% confluency were scraped off and then lysed in a complete radioimmunoprecipitation assay lysis buffer. A total of 100 μl of the cell extract were incubated with anti-DKC1 or control IgG (Millipore)-conjugated magnetic beads at 4°C for 6 h, and anti-IgG was used as a negative control. The beads were then washed with a washing buffer and the complexes were incubated with Proteinase K for 30 min at 55°C to remove the protein. Finally, immunoprecipitated RNA was purified and analyzed by RT-qPCR.
RNA pulldown assay
As described previously [Citation33], biotin-labeled FLT1P1 and its antisense RNA were transcribed in vitro based on the corresponding PCR product using T7 RNA polymerase (Ambio Life) and Biotin RNA Labeling Mix (Roche, Mannheim, Germany). The obtained product was purified using RNeasy Plus Mini Kit (Qiagen). For the pulldown assay, 20 µl Dynabeads M-280 Streptavidin beads (Thermo Fisher Scientific, MA, USA) were activated and blocked with 10 µg/ml RNase-free BSA and yeast tRNA (Sigma) for 30 min at 4°C. Cell lysates were incubated with the beads at room temperature for 2 h, followed by pulldown, RNA extraction, and subsequent RT-qPCR quantification.
RNA stability assay
For detecting FLT1 mRNA stability, trophoblast cells were treated with 10 µg/ml actinomycin D (Sigma) after transfecting with sh-FLT1P1 or sh-DKC1 [Citation34]. At 3 h, 6 h, and 9 h, cells were collected, and RNA extraction was performed by using TRIzol reagent (Invitrogen). RT-qPCR was used to measure the FLT1 mRNA level.
Statistical analysis
Statistical analysis was performed using Graphpad Prism 5 software (GraphPad, USA), and the results are presented as the mean ± standard deviation (SD). Each assay was repeated in triplicate. A comparison between groups was performed using Student’s t test or one-way or two-way analysis of variance (ANOVA). P value less than 0.05 was considered statistically significant.
Results
In this study, we focused on identifying the role of the pseudogene FLT1P1 in trophoblast cell proliferation and angiogenesis and mechanistically analyzing how FLT1P1 exerts its function in PE. We hypothesized that FLT1P1 may exert its function in PE by interacting with the RBP DKC1 to stabilize its cognate gene FLT1. Our results showed that FLT1P1 and FLT1 play a vital role in PE by regulating trophoblast cell proliferation and angiogenesis. Moreover, FLT1P1 increases FLT1 mRNA stability via recruiting DKC1.
FLT1P1 knockdown promotes the proliferation and angiogenesis in trophoblast cells
Human PTBs were isolated from PE and healthy placentas. The results of RT-qPCR showed that FLT1P1 was upregulated in PTBs isolated from PE patients compared with healthy controls (Fig. S1A). Next, we investigated the biological relevance of FLT1P1 in PE progression using two trophoblast cell line (HTR8 and BeWo). As shown in , the expression of FLT1P1 in HTR8 and BeWo cells was effectively knocked down by transfection with the specific shRNA (sh-FLT1P1) compared to the sh-NC. Next, the functional role of FLT1P1 in trophoblast cell proliferation and angiogenesis was assessed by CCK-8 assay, tube formation assay, and western blot analysis. According to CCK-8 assay, the proliferative ability of trophoblast cells was increased after knocking down FLT1P1 (). The results of the tube formation assay showed that FLT1P1 downregulation significantly increased the number of junctions and nodes in HTR8 and BeWo cells (). Furthermore, the expression levels of angiogenesis-associated markers (VEGFA, FGF2 and TGF-β) was evaluated using western blot. As shown in , the expression levels of VEGFA, FGF2, and TGF-β were all increased in the presence of sh-FLT1P1 (). Overall, these findings showed that FLT1P1 knockdown increases the proliferation and angiogenesis in trophoblast cells.
Figure 1. FLT1P1 knockdown promotes trophoblast cell proliferation and angiogenesis. (a) FLT1P1 expression in trophoblast cells transfected with sh-FLT1P1 or sh-NC was measured by RT-qPCR. (b) The proliferation of HTR8 and BeWo cells after FLT1P1 knockdown was detected by CCK-8 assay. (c) The angiogenesis in sh-FLT1P1-transfected HTR8 and BeWo cells was assessed by tube formation assay. (d) Western blot analysis was performed to measure the VEGFA, FGF2 and TGF-β protein levels in HTR8 and BeWo cells transfected with sh-FLT1P1 or sh-NC. *p < 0.05, ***p < 0.001
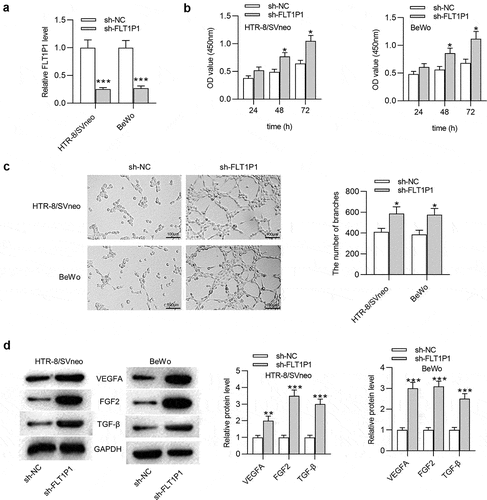
FLT1 knockdown promotes trophoblast cell proliferation and angiogenesis
As the cognate gene of FLT1P1, the biological role of FLT1 in trophoblast cells was investigated. The results of RT-qPCR and western blot showed that the FLT1 mRNA and protein levels were upregulated in PTBs isolated from PE patients compared with healthy controls (Fig. S1B-1 C). The sh-FLT1 was transfected into HTR8 and BeWo cells to silence FLT1. The results of RT-qPCR showed that FLT1 expression was significantly downregulated in the sh-FLT1-transfetced HTR8 and BeWo cells (). CCK-8 assay manifested that sh-FLT1 transfection caused an increase in cell proliferation (). Next, the angiogenesis ability after sh-FLT1 transfection was evaluated by tube formation assay. The results demonstrated that the angiogenesis in HTR8 and BeWo cells was promoted after silencing FLT1 (). Additionally, the results of the western blot indicated that FLT1 knockdown contributed to elevated protein levels of VEGFA, FGF2 and TGF-β in HTR8 and BeWo cells (). In conclusion, FLT1 knockdown increases trophoblast cell proliferation and angiogenesis.
Figure 2. FLT1 knockdown promotes the proliferation and angiogenesis in trophoblast cells. (a) FLT1 expression in trophoblast cells transfected with sh-FLT1 or sh-NC was measured by RT-qPCR. (b) CCK-8 assay was performed to assess the proliferation in sh-FLT1-transfected HTR8 and BeWo cells. (c) The angiogenesis in HTR8 and BeWo cells after FLT1 knockdown was evaluated by tube formation assay. (d) The VEGFA, FGF2 and TGF-β protein levels in sh-FLT1-transfected HTR8 and BeWo cells were measured by western blot analysis. *p < 0.05, **p < 0.01, ***p < 0.001
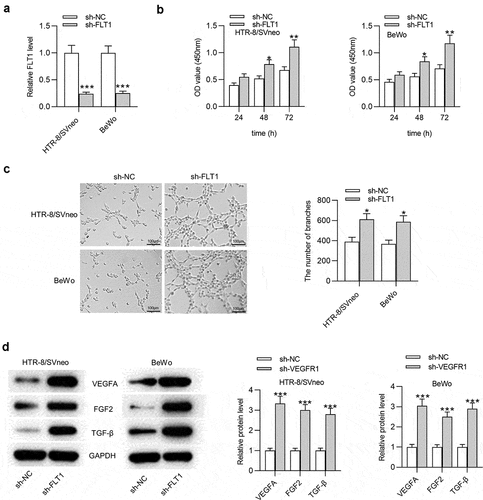
DKC1 acts as an RBP for FLT1P1 and FLT1
Next, we investigated whether there is a regulatory mechanism between FLT1P1 and FLT1. Evidence has confirmed that the pseudogene could recruit RBPs to modulate its target gene expression [Citation18,Citation19]. Thus, we sought to identify the common RBP for FLT1P1 and FLT1. At the starBase website (http://starbase.sysu.edu.cn/), DKC1 was predicted as an RBP that has the potential to interact with FLT1P1 and FLT1 (). Then, the binding capacity between FLT1P1 (or FLT1) and DKC1 was further predicted at the RPISeq website (http://pridb.gdcb.iastate.edu/). The score of RF Classifier is over 0.5 and the score of SVM Classifier is over 0.8, indicating that FLT1P1 (or FLT1) stands a good chance of binding to DKC1 (). As predicted in starBase, the motif of DKC1 in FLT1P1 and FLT1 was obtained (). RIP assay showed that FLT1P1 and FLT1 were both enriched in the beads conjugated with anti-DKC1 compared to anti-IgG in trophoblast cells (). To further investigate the interaction between FLT1P1 (or FLT1) and DKC1, a biotin-labeled FLT1P1 (or FLT1) probe was used to perform RNA pulldown assay in HTR8 and BeWo cells. The results revealed that the DKC1 was significantly enriched in the FLT1P1 (or FLT1) sense probe compared with the sense probe (). These results suggested that FLT1P1 (or FLT1) could bind to DKC1.
Figure 3. The interaction between DKC1 and FLT1P1 (or FLT1). (a) The predicted RBP for FLT1P1 and FLT1 was obtained in starBase. (b) The prediction of the interaction probabilities of FLT1P1 or FLT1 with DKC1 by RPISeq (http://pridb.gdcb.iastate.edu/RPISeq/). Predictions with probabilities > 0.5 are considered ‘positive’, indicating that the corresponding RNA and protein are likely to interact. (c) The motif of DKC1 in FLT1P1 and FLT1. (d-e) The binding of DKC1 to FLT1P1 (or FLT1) was validated by RIP and RNA pull down assays. ***p < 0.001
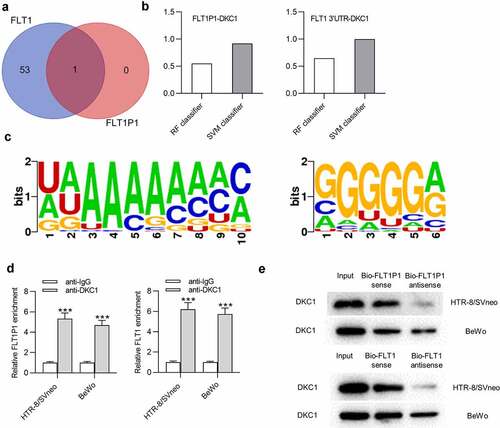
FLT1P1 increases FLT1 mRNA stability by recruiting DKC1
Subsequently, to test whether FLT1P1/DKC1 exerts function on FLT1 mRNA stability, we tested FLT1 mRNA expression with the treatment of Actinomycin D. The results showed that FLT1P1 knockdown decreased FLT1 mRNA expression under Actinomycin D treatment (). We then silenced DKC1 expression with the transfection of sh-DKC1 in trophoblast cells for further analysis (). As shown, FLT1 mRNA stability was significantly decreased by DKC1 downregulation in HTR8 and BeWo cells treated with actinomycin D (). Furthermore, we found that the FLT1 protein level in cells was also reduced by DKC1 downregulation (). Overall, FLT1P1 maintains FLT1 mRNA stability via recruiting DKC1.
Figure 4. FLT1P1 increases FLT1 mRNA stability by recruiting DKC1. (a) FLT1 mRNA expression under actinomycin D treatment in the sh-FLT1P1 and sh-NC groups was measured by RT-qPCR. (b) The transfection efficiency of sh-DKC1 was assessed by RT-qPCR. (c) FLT1 mRNA expression under actinomycin D treatment in the sh-DKC1 and sh-NC groups was measured by RT-qPCR. (d) The FLT1 protein level in trophoblast cells transfected with sh-DKC1 was measured by western blot analysis. *p < 0.05, **p < 0.01, ***p < 0.001
FLT1P1 regulates trophoblast cell proliferation and angiogenesis by mediating FLT1
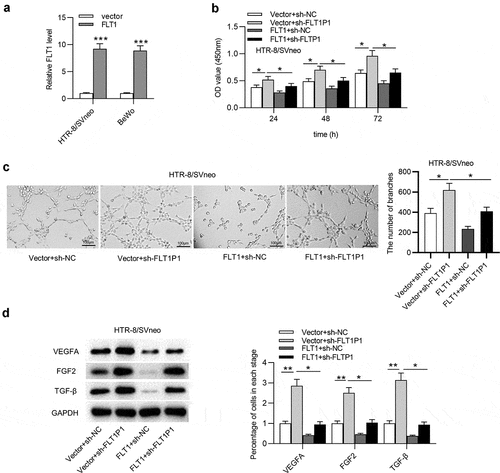
Finally, we verified whether FLT1P1 regulates trophoblast cells by FLT1, and rescue assays were performed. The overexpression efficiency of FLT1 was evaluated by RT-qPCR. The results showed that FLT1 was effectively overexpressed with pcDNA3.1/FLT1 transfection in HTR8 and BeWo cells (). As shown in CCK-8 assay, FLT1 overexpression attenuated the promotive effect of FLT1P1 knockdown on trophoblast cell proliferation (). We further discovered that angiogenesis restored by FLT1P1 knockdown was inhibited by FLT1 overexpression (). In addition, the VEGFA, FGF2, and TGF-β protein levels increased by FLT1P1 knockdown was reduced after FLT1 overexpression (). Collectively, FLT1P1 upregulates FLT1 expression to inhibit trophoblast cell proliferation and angiogenesis.
Figure 5. FLT1P1 increases FLT1 expression to regulate trophoblast cell proliferation and angiogenesis. (a) The overexpression efficiency of FLT1 was confirmed by RT-qPCR analysis. (b) The proliferative ability of trophoblast cells in each group was measured by CCK-8 assay. (c-d) Angiogenesis and angiogenesis-relevant protein levels were evaluated by tube formation assay and western blot analysis. *p < 0.05, **p < 0.01, ***p < 0.001
Discussion
Pseudogenes, which are fairly common (~0.7% of DNA sequence) in the human genome, as well as lncRNAs are traditionally claimed to not yield functional mRNAs and not translated into proteins consequently, thus regarded as garbage fragments or dark matter in the genome [Citation35]; however, evidence in recent years has indicated that pseudogene regulates various aspects of cell biology, and there is an increasing attention on its potential contribution to disease cause [Citation36]. A previous study showed that the newly identified pseudogenes BNIP3P1, HK2P1, and PGK1P1 that encode lncRNA are key PE-related genes [Citation37]. Hexokinase 2 pseudogene 1 (HK2P10) expression is correlated with abnormal decidualization and might lead to the occurrence of PE [Citation38]. However, the related mechanisms and the roles of abnormally expressed pseudogenes in PE have not been functionally characterized to date. Therefore, the identification of key pseudogenes associated with PE is critical to identifying novel therapeutic targets. A study pointed out that FLT1P1 is expressed at a high level in preeclamptic placentas [Citation26], implying that pseudogene FLT1P1 might play an important role in the development of PE. The biological function of FLT1P1 has not been investigated previously. In this study, we found that FLT1P1 was upregulated in PTBs isolated from PE patients compared with healthy controls. The functional assays suggested that the downregulation of FLT1P1 altered the proliferative capacity of HTR8 and BeWo cells. Defects in decidualization cause inadequate placentation and angiogenesis, which could give rise to PE [Citation39]. VEGF is famous as an angiogenesis factor in many tissues and its decreased expression has been implicated in the pathophysiology of PE [Citation40,Citation41]. Here, it was demonstrated that FLT1P1 knockdown promoted the angiogenesis in the trophoblast, demonstrating the key role of FLT1P1 in PE.
Pseudogenes have a high sequence similarity to their parental protein-coding genes, which generates the potential for sequence-specific regulation [Citation36]. The pseudogene FLT1P1 shares molecular ancestry with the cognate gene FLT1 (VEGFR1) in humans and high primates [Citation25]. The VEGF family has been implicated as an important regulator of blood vessel formation in both health and disease states, including PE, tumor neovascularization, and diabetic retinopathy [Citation42]. FLT1 (VEGFR1) is a type V protein-tyrosine kinase receptor that is crucial for cell proliferation and differentiation and is expressed in vascular endothelial cells, placental trophoblast cells, and peripheral blood monocytes. A soluble form of FLT1 is markedly increased during the last 2 months of gestation in those with PE compared with normotensive pregnant controls, which is involved in the endothelial dysfunction characterizing the pregnancy disorder of PE [Citation43]. Here, our study demonstrated that FLT1 was upregulated in PTBs isolated from PE patients. FLT1 knockdown promoted the proliferation and angiogenesis in the trophoblast. One of the most commonly described biological feature of processed pseudogenes is the ability to influence the expression of their parental coding genes. The high sequence similarity between these RNA pairs sets up a certain level of competition for posttranscriptional regulators, including, among others, RBPs [Citation44]. Thus, we further explored the regulatory relationship between FLT1P1 and FLT1. Through bioinformatics analysis, DKC1 was predicted to be an RBP interacting with both FLT1P1 and FLT1. DKC1 is a key RBP encoding a protein responsible for telomerase holoenzyme complex stability [Citation45]. In our study, we confirmed that DKC1 could bind to either FLT1P1 or FLT1. Moreover, the knockdown of FLT1P1 or DKC1 effectively decreased the mRNA stability and protein level of FLT1. Therefore, we demonstrated that FLT1P1 increases FLT1 mRNA stability by recruiting DKC1 in this study.
Conclusion
In conclusion, we demonstrated that FLT1P1 and FLT1 play a vital role in PE by regulating trophoblast cell proliferation and angiogenesis. Moreover, FLT1P1 increases FLT1 mRNA stability via recruiting DKC1. These results show that dysregulated FLT1P1 and FLT1 may be related to the occurrence of PE, suggesting that FLT1P1 and FLT1 could act as useful biomarkers for the diagnosis of PE. The present study is not without limitations. First, clinical samples need to be collected from PE patients to further verify the clinical significance of our findings. Second, the related signaling pathways targeted by the FLT1P1/FLT1remain unclear and require further investigations.
Supplemental Material
Download ()Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Ramos JGL, Sass N, Costa SHM. Preeclampsia. Rev Bras Ginecol Obstet. 2017 Sep;39(9):496–512.
- Murthi P, Pinar AA, Dimitriadis E, et al.Inflammasomes-A Molecular Link for Altered Immunoregulation and Inflammation Mediated Vascular Dysfunction in Preeclampsia. Int J Mol Sci. 2020 Feb 19;Vol. 21:4.
- Sánchez-Aranguren LC, Prada CE, Riaño-Medina CE, et al. Endothelial dysfunction and preeclampsia: role of oxidative stress. Front Physiol. 2014;5:372.
- Phipps EA, Thadhani R, Benzing T, et al. Pre-eclampsia: pathogenesis, novel diagnostics and therapies. Nat Rev Nephrol. 2019 May;15(5):275–289.
- Rana S, Lemoine E, Granger JP, et al. Preeclampsia: pathophysiology, challenges, and perspectives. Circ Res. 2019 Mar 29;124(7):1094–1112.
- Shen Y, Liu S, Fan J, et al. Nuclear retention of the lncRNA SNHG1 by doxorubicin attenuates hnRNPC-p53 protein interactions. EMBO Rep. 2017 Apr;18(4):536–548.
- Chen X, Yan CC, Zhang X, et al. Long non-coding RNAs and complex diseases: from experimental results to computational models. Brief Bioinform. 2017 Jul 1;18(4):558–576.
- Xu Y, Ge Z, Zhang E, et al. The lncRNA TUG1 modulates proliferation in trophoblast cells via epigenetic suppression of RND3. Cell Death Dis. 2017 Oct 12;8(10):e3104.
- Cao C, Li J, Li J, et al. Long non-coding RNA Uc.187 Is upregulated in preeclampsia and modulates proliferation, apoptosis, and invasion of HTR-8/SVneo trophoblast cells. J Cell Biochem. 2017 Jun;118(6):1462–1470.
- Zhang Y, Zou Y, Wang W, et al. Down-regulated long non-coding RNA MEG3 and its effect on promoting apoptosis and suppressing migration of trophoblast cells. J Cell Biochem. 2015 Apr;116(4):542–550.
- Chen H, Meng T, Liu X, et al. Long non-coding RNA MALAT-1 is downregulated in preeclampsia and regulates proliferation, apoptosis, migration and invasion of JEG-3 trophoblast cells. Int J Clin Exp Pathol. 2015;8(10):12718–12727.
- Karreth FA, Reschke M, Ruocco A, et al. The BRAF pseudogene functions as a competitive endogenous RNA and induces lymphoma in vivo. Cell. 2015 Apr 9;161(2):319–332.
- Lai Y, Li J, Zhong L, et al. The pseudogene PTENP1 regulates smooth muscle cells as a competing endogenous RNA. Clin Sci (Lond). 2019 Jul 15;133(13):1439–1455.
- Zhou LY, Zhai LL, Yin JY, et al. Pseudogene BMI1P1 expression as a novel predictor for acute myeloid leukemia development and prognosis. Oncotarget. 2016 Jul 26;7(30):47376–47386.
- Gupta A, Brown CT, Zheng YH, et al. Differentially-Expressed pseudogenes in HIV-1 infection. Viruses. 2015 Sep 29;7(10):5191–5205.
- Yu L, Kuang LY, He F, et al. The role and molecular mechanism of long noncoding RNA-MEG3 in the pathogenesis of preeclampsia. Reprod Sci. 2018 Dec;25(12):1619–1628.
- Tong J, Yang J, Lv H, et al. Dysfunction of pseudogene PGK1P2 is involved in preeclampsia by acting as a competing endogenous RNA of PGK1. Pregnancy Hypertens. 2018 Jul;13:37–45.
- D’Angelo D, Arra C, Fusco A. RPSAP52 lncRNA inhibits p21Waf1/CIP expression by interacting with the RNA binding protein HuR. Oncol Res. 2020 Mar 27;28(2):191–201.
- Oliveira-Mateos C, Sánchez-Castillo A, Soler M, et al. The transcribed pseudogene RPSAP52 enhances the oncofetal HMGA2-IGF2BP2-RAS axis through LIN28B-dependent and independent let-7 inhibition. Nat Commun. 2019 Sep 4;10(1):3979.
- Heiss NS, Knight SW, Vulliamy TJ, et al. X-linked dyskeratosis congenita is caused by mutations in a highly conserved gene with putative nucleolar functions. Nat Genet. 1998 May;19(1):32–38.
- Mitchell JR, Wood E, Collins K. A telomerase component is defective in the human disease dyskeratosis congenita. Nature. 1999 Dec 2;402(6761):551–555.
- Yang Z, Wang Z, LncRNA DY. MEG3 inhibits non-small cell lung cancer via interaction with DKC1 protein. Oncol Lett. 2020 Sep;20(3):2183–2190.
- Wang R, Zou L. Downregulation of LncRNA-MEG3 promotes HTR8/SVneo cells apoptosis and attenuates its migration by repressing Notch1 signal in preeclampsia. Reproduction. 2020 Jul;160(1):21–29.
- Zhang J, Liu X, Gao Y. The long noncoding RNA MEG3 regulates Ras-MAPK pathway through RASA1 in trophoblast and is associated with unexplained recurrent spontaneous abortion. Mol Med. 2021 Jul 8;27(1):70.
- Ye X, Fan F, Bhattacharya R, et al. VEGFR-1 Pseudogene expression and regulatory function in human colorectal cancer cells. Mol Cancer Res. 2015 Sep;13(9):1274–1282.
- Song X, Luo X, Gao Q, et al. Dysregulation of LncRNAs in placenta and pathogenesis of preeclampsia. Curr Drug Targets. 2017;18(10):1165–1170.
- Sagrillo-Fagundes L, Clabault H, Laurent L, et al. Human primary trophoblast cell culture model to study the protective effects of melatonin against hypoxia/reoxygenation-induced disruption. J Vis Exp. 2016;30:113.
- Zou Z, Ma T, He X, et al. Long intergenic non-coding RNA 00324 promotes gastric cancer cell proliferation via binding with HuR and stabilizing FAM83B expression. Cell Death Dis. 2018 Jun 18;9(7):717.
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001 Dec;25(4):402–408.
- Li P, Gu L, Bian Q, et al. Long non-coding RNA MALAT1 enhances the protective effect of dexmedetomidine on acute lung injury by sponging miR-135a-5p to downregulate the ratio of X-box binding proteins XBP-1S/XBP-1U. Bioengineered. 2021 Dec;12(1):6377–6389.
- Ritter A, Roth S, Kreis NN, et al. Primary cilia in trophoblastic cells: potential involvement in preeclampsia. Hypertension. 2020 Nov;76(5):1491–1505.
- Chen L, Fang Z, Wang X, et al. G protein-coupled receptor 39 activation alleviates oxidized low-density lipoprotein-induced macrophage inflammatory response, lipid accumulation and apoptosis by inducing A20 expression. Bioengineered. 2021;Dec;12(1):4070–4080.
- Yuan S, Si W, Zhuang K, et al. LncRNA UCID Promotes hepatocellular carcinoma metastasis via stabilization of snail. Onco Targets Ther. 2021;14:725–736.
- Barros-Silva D, Lobo J, Guimarães-Teixeira C, et al. VIRMA-Dependent N6-methyladenosine modifications regulate the expression of long non-coding RNAs CCAT1 and CCAT2 in prostate cancer. Cancers (Basel). 2020 Mar 25;12(4):4.
- Palazzo AF, Lee ES. Non-coding RNA: what is functional and what is junk? Front Genet. 2015;6:2.
- Milligan MJ, Lipovich L. Pseudogene-derived lncRNAs: emerging regulators of gene expression. Front Genet. 2014;5:476.
- Tong J, Zhao W, Lv H, et al. Transcriptomic profiling in human decidua of severe preeclampsia detected by RNA sequencing. J Cell Biochem. 2018 Jan;119(1):607–615.
- Lv H, Tong J, Yang J, et al. Dysregulated pseudogene HK2P1 may contribute to preeclampsia as a competing endogenous RNA for hexokinase 2 by impairing decidualization. Hypertension. 2018 Apr;71(4):648–658.
- Cha J, Sun X, Dey SK. Mechanisms of implantation: strategies for successful pregnancy. Nat Med. 2012 Dec;18(12):1754–1767.
- Zhang D, Chang X, Bai J, et al. The study of cyclooxygenase 2 in human decidua of preeclampsia. Biol Reprod. 2016 Sep;95(3):56.
- Masoura S, Kalogiannidis I, Makedou K, et al. Biomarkers of endothelial dysfunction in preeclampsia and neonatal morbidity: a case–control study. Eur J Obstet Gynecol Reprod Biol. 2014 Apr;175:119–123.
- Amosco MD, Villar VA, Naniong JM, et al. VEGF-A and VEGFR1 SNPs associate with preeclampsia in a Philippine population. Clin Exp Hypertens. 2016;38(7):578–585.
- Levine RJ, Maynard SE, Qian C, et al. Circulating angiogenic factors and the risk of preeclampsia. N Engl J Med. 2004 Feb 12;350(7):672–683.
- Chiefari E, Arcidiacono B, Mirabelli M, et al. Methods to study protein-binding to pseudogene transcripts. Methods Mol Biol. 2021;2324:187–202.
- Donaires FS, Alves-Paiva RM, Gutierrez-Rodrigues F, et al. Telomere dynamics and hematopoietic differentiation of human DKC1-mutant induced pluripotent stem cells. Stem Cell Res. 2019 Oct;40:101540.
