ABSTRACT
Colonic enterocytes form a rapidly renewing epithelium and barrier to luminal antigens. During renewal, coordinated expression of the claudin family of genes is vital to maintain the epithelial barrier. Disruption of this process contributes to barrier compromise and mucosal inflammatory diseases. However, little is known about the regulation of this critical aspect of epithelial cell differentiation. In order to identify claudin regulatory factors we utilized high-throughput gene microarrays and correlation analyses. We identified complex expression gradients for the transcription factors Hopx, Hnf4a, Klf4 and Tcf7l2, as well as 12 claudins, during differentiation. In vitro confirmatory methods identified 2 pathways that stimulate claudin expression; Hopx/Klf4 activation of Cldn4, 7 and 15, and Tcf7l2/Hnf4a up-regulation of Cldn23. Chromatin immunoprecipitation confirmed a Tcf7l2/Hnf4a/Claudin23 cascade. Furthermore, Hnf4a conditional knockout mice fail to induce Cldn23 during colonocyte differentiation. In conclusion, we report a comprehensive screen of colonic claudin gene expression and discover spatiotemporal Hopx/Klf4 and Tcf7l2/Hnf4a signaling as stimulators of colonic epithelial barrier differentiation.
KEYWORDS:
Introduction
One important aspect of colonic epithelial function is to provide a selective barrier between the body and the gut lumen. Colonic barrier function requires that epithelial cells within the tissue maintain cell-to-cell contact structures called Tight Junctions. Tight Junctions (TJs) are multi-protein complexes containing 3 main transmembrane protein families: claudins, occludin, and Junctional Adhesion Molecules (JAMs).Citation1 Of the 3, claudins (Cldn) constitute the paracellular seal by spanning the extracellular space between cells.Citation2,3 Importantly, aberrant claudin gene expression is thought to contribute to diseases such as colon cancer and inflammatory diseases.Citation4-6 Therefore, there is a need to further determine factors and mechanisms that regulate claudin gene expression in health and disease.
The claudin-based TJ seal is semi-permeable to the passage of small molecules and ions, and the physiological properties of the TJ are determined by differential claudin gene expression.Citation3,7 Indeed, claudin expression varies among tissue types and plays crucial roles in wide ranging biological processes: from ion resorption in the kidney to glucose transport in the intestine.Citation8,9 There are 24 claudin gene family members in mice, with varied expression patterns along the gastrointestinal tract.Citation10 Differential claudin expression within tissues is less well documented, but colonic tissues have been reported to express at least claudins 1–4, 7, 8, 10 and 15 (reviewed in Citation11). These claudins are under complex spatial regulation, with specific claudin genes expressed within colonic crypt-base regions (claudins 2 and 10) and others expressed in the lumen-facing surface epithelial cell populations (claudins 3, 4, and 7).
The colonic epithelial lining is highly regenerative, with resident stem cells replenishing the tissue over several days.Citation12,13 Epithelial cells are generated from a proliferative stem cell compartment within the colonic crypt-base. Progenitor cells divide and migrate toward the surface-cell region of the gut to be ultimately shed into the lumen. Under physiological conditions, epithelial tissue homeostasis is thought to involve concerted cellular signaling and transcription factor cascades that control epithelial homeostasis; encompassing proliferation, differentiation and cell death. Comparative gene expression studies have identified signaling molecules including bone morphogenic protein antagonists, Notch, Wnt, Ephrin, and Myc signaling pathways, as important regulators of intestinal proliferation and differentiation.Citation14-17 Studies that describe the specific claudin regulation in the colon are rare. In one study, claudin 7 was found to be suppressed in colonic crypt-base regions by Tcf-4/Sox9 signaling.Citation18 Furthermore, claudins 1 and 2 were shown to be regulated by Cdx-2/GATA-4 in undifferentiated intestinal cells.Citation19,20 Claudins are a large gene family whose contribution to barrier function is determined by the complex interactions of multiple family members. However, little is known concerning factors that regulate spatiotemporal claudin differentiation within the intestinal crypt.
In this study, we investigated claudin gene regulation in the colon by combining high-throughput gene microarrays, gene expression correlation analyses, and combined in vitro and in vivo confirmatory methods. We first hypothesized that the transcription factors responsible for claudin regulation would correlate with claudin gene expression along the crypt-base-to-surface cell axis. Therefore, we performed comparative gene expression studies from colonic IECs collected by laser capture microdissection (LCM). Cells were collected from 2 spatially distinct regions sequentially (i.e., distal colon crypt-base and luminal surface cells) and high-throughput analysis of gene microarray experiments (using the Illumina platform) identified transcriptome variation between cellular subpopulations along the crypt-base-to-surface axis. Nine claudins (Cldn2-5, 7, 9, 10, 12, and 23) and 3 transcription factors (Hopx, Tcf7l2, and Hnf4a) were selected for further analysis, and gradated expression of these genes was subsequently validated by real-time PCR. Claudin transcript abundance was then determined by quantitative ddPCR, indicating 12 claudin genes are abundantly expressed in colonic crypts. Ectopic expression of the transcription factors Hopx, Hnf4a and Tcf7l2 in vitro confirmed their role in claudin regulation. Under these conditions, Hopx was found to stimulate Cldn4, 7, 15 and Klf4, while Tcf7l2 was found to stimulate multiple claudins and Hnf4a expression. Hnf4a stimulated the expression of a singular claudin, Cldn23. Binding of Tcf7l2 to the Hnf4a gene promoter, as well as HNF4a binding to the Cldn23 promoter, was then determined by Chromatin Immunoprecipitation (ChIP). Moreover, in Hnf4a conditional knockout mice, Cldn23 mRNA levels were decreased significantly, as these mice failed to induce Cldn23 in surface cell populations.
Results
Gene ontology analysis of differentially regulated genes along the crypt-base–surface axis of the mouse distal colon
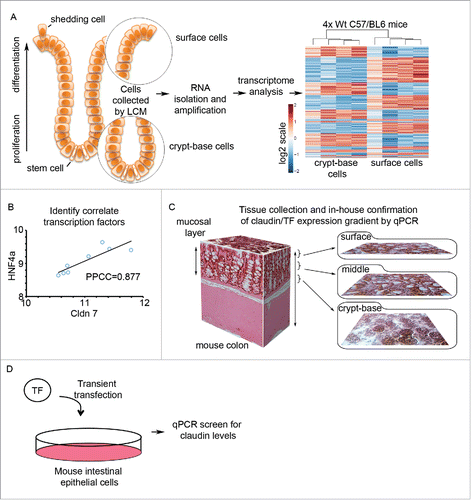
Claudin gene expression is responsible for the variance of paracellular barrier properties among epithelial and endothelial tissues. Interestingly, claudin expression varies dramatically within tissues as well, as exemplified by claudin expression within colonic crypts. We sought to investigate factors that regulate claudin gene expression occurring along the colonic IEC crypt-base to surface axis (). We therefore isolated these IEC populations using laser capture microdissection (LCM) from 2 spatially distinct regions of the colon. Four male C57/BL6 (wild-type) mice were sacrificed and dissected and the distal colon was removed. Tissue segments were then processed such that both crypt-base and surface cell population are evident for each crypt (). Individual crypts were then sequentially microdissected using LCM, first removing the crypt-base cells, and then harvesting the surface cells. The resulting samples were processed for RNA isolation, cDNA generation, and amplification (see Methods and Materials). Whole genome expression was assessed for each sample using the Illumina MouseWG-6_v1.1 platform and post-processed using the default settings of Illumina Genome Studio software. Microarray data expression analysis (see Materials and Methods for the detailed Microarray Analysis) showed 2,639 probe sets to be statistically significantly differentially expressed between these 2 crypt compartments with stringent statistical significance criteria (False Discovery Rate (FDR) equal to 0.2). Pathway enrichment was then performed on the 2,639 differentially expressed probes using GeneGO (2,450 unique genes), which showed that 18 out of 20 most enriched pathways of statistical significance (FDR ≤ 2.307e-5) were directly or indirectly related to cell cycle and cell adhesion processes (Table S2, top 10 are shown). Thus, we found that the majority of the most significant pathways responsible for differential gene expression of the crypt base vs. the surface epithelial cells were associated with cell adhesion and cell cycle. contains an illustration and examples of workflow and data analysis. As shown in , transcription factor/claudin correlation analysis was performed on differentially expressed genes to identify potential positive regulators of claudin gene expression. For example, Hnf4a signal levels within the array positively correlate with claudin 7 (Pearson's correlation coefficient r = 0.88, at p-value = 0.004). In , differentially regulated claudin genes, as well as correlated transcription factors, were confirmed by qPCR using tissue collected by cryosectioning method as illustrated (as described in Citation21). Like LCM, crypt cryosectioning preserves the native cellular environment until RNA collection and is a high yield method that does not require RNA amplification.
Figure 1. Transcriptome analysis of spatially distinct epithelial cell populations in mouse colon. (A) Schematic of the colonic epithelial monolayer lining the gut. Two cell regions were compared, crypt-base cell populations containing the proliferative compartment and differentiated surface cells. Cells were extracted sequentially using laser capture microscopy (LCM, n = 4/sample). RNA was then converted to cDNA and amplified. Subsequent microarray analysis identified spatially distinct gene expression patterns between surface and crypt-base cell populations. (B) Perform Pearson Product-Moment Correlation Coefficient (PPCC) to identify transcription factors that correlate with claudin gene expression. (C) Schematic of colonic mucosa sectioning method for subsequent confirmation of graded gene expression. (D) Transcription factors of interest were overexpressed in mouse intestinal cells, which were subsequently harvested for gene expression analysis and chromatin immunoprecipitation (ChIP).
Differentially expressed claudins correlate with specific transcription factor expression
From the GeneGO analysis, the most significant cellular processes identified in the regulation of intestinal epithelial cell (IEC) differentiation were Cell Cycle and Cell Adhesion (Table S2). For the purposes of this study, we focus on cell adhesion genes, particularly the genes for claudin family proteins (). A total of 6 claudin probes (designed to hybridize Cldn2, Cldn5, Cldn7, Cldn3, Cldn4 and Cldn23) were differentially expressed along the crypt-base-to-surface axis, 4 claudin probes (designed to hybridize Cldn9, Cldn10, Cldn12 and Cldn19) were detected but did not change expression, and the remaining 17 claudin probes on the array were detected in the crypt-base compartment only (Cldn15) or they were not detected at all based on the p-values of signal detection per sample (see Table S3). Finally, the Tight Junction genes encoding ZO-1 and ZO-2 (Tjp1 and Tjp2) and occludin (Ocln) changed expression significantly, all showing enrichment in the surface cell population (). The gene expression differences of the 6 claudin genes were able to distinguish the surface from the crypt-base compartments according to additional clustering analyses ()
Figure 2. Tight Junction related cell adhesion genes are heavily represented among the differentially expressed genes. (A) Tight junction genes identified as differentially expressed by microarray analysis. Genes colored in red indicate crypt-base enrichment, blue text indicates surface cell enrichment. (B) Hierarchical clustering of the 6 differentially expressed claudins. (C) Claudin genes detected that did not exhibit expression changes. (D) Transcription factors were identified for further analysis (red, Hopx and Tcf7l2) based on fold change and positive Cldn2 Pearson Product-Moment Correlation Coefficient (PPCC). (E) Transcription factors significantly differentially expressed, with red indicating higher crypt-base expression. (F) Hierarchical clustering of 4 differentially expressed transcription factors in the crypt compared to the surface.
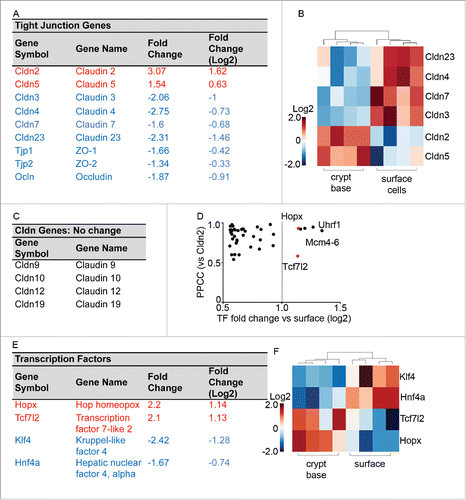
In order to identify candidate transcription factors (TFs) that regulate claudins during IEC differentiation we screened our dataset for TFs, by combining 2 online databases (RIKEN database and Animal transcription factor database). From the combined 2,301 transcription factors within these databases, 192 were found to be differentially expressed (using a 2-sample, 2-tail, unequal variance t-test with FDR<0.2) between the crypt-base and surface cell populations, 69 of these exhibited a change more than 1.5-fold (fold change > 0.6 in the log2 scale, Table S3). We then focused on 2 highly up-regulated TFs in crypt-base populations which correlated positively with crypt-base expressed Cldns: Hopx and Tcf7l2 (). Uhrf1 and Mcm 4, 5, and 6 also had high correlation coefficients and differential expression, yet were not pursued further as they do not act as transcription factors (PPCC vs. Cldn2, ). We also investigated the TFs Hnf4a and Klf4, as their expression was found to be enhanced in differentiated surface epithelia (). Importantly, Hnf4a has been shown to play a role in claudin expression.Citation22,23 In summary, specific changes in claudin gene expression correlate with changes in the expression of the TFs: Hnf4a, Hopx, and Tcf7l2.
Real-time PCR validation of array findings
Based on the above analysis, we focused on 6 claudin genes and 4 transcription factors of interest that were found to be differentially expressed along the crypt-base-to-surface cell axis. In order to validate these observations, we confirmed the gene expression patterns using real-time PCR (qPCR). We extracted spatially distinct IEC cell populations using a serial cryosectioning method, with sequential 10 μm sections pooled into crypt-base, middle, and surface epithelial cell samples (,Citation21). As shown in , real-time PCR analysis was used to confirm a crypt-base enrichment for Cldn2, and 5, as well as surface enrichment for Cldn3, 4, and 23. Cldn7 and Cldn9 levels exhibit a trend toward enrichment in surface or crypt cell populations respectively, yet these trends failed to reach statistical significance. Similar to our microarray findings, Cldn10 and 12 mRNA varied little between the different subpopulations. mRNA levels of the Tight-Junction genes encoding Ocln, and Tjp1 did not change between the crypt-base and the surface with a small but significant enrichment in Tjp2 observed in the crypt-base region (). We next confirmed the differential expression of transcription factors Hopx, Hnf4a, and Klf4 in the crypt-base-surface axis were consistent with the microarray results (). As a control for sample reliability, we monitored Klf4 and Klf5 expression, as these factors are documented to exhibit an inverse expression gradient along the crypt-surface axis.Citation24,25 Importantly, Tcf7l2 exhibited surface region enrichment; therefore we are unable to confirm graded expression of this gene product. In summary, the PCR results corroborated the array findings suggesting differential expression of claudin genes in the crypt-to-surface axis correlated with changes in specific TFs.
Figure 3. Confirmation of microarray results by real-time PCR. (A) Relative mRNA expression of claudin genes along the crypt-surface axis normalized to crypt values. (B) Relative mRNA expression of Tight Junction related genes. C. Relative mRNA expression of candidate regulatory transcription factors (A, B and C. *p < 0.05, **p < 0.01, one-way ANOVA, surface vs. crypt-base). (D) ddPCR showing copies of claudin transcript in surface cell populations, (E) transitional middle cells, (F) and crypt-base cells (expression normalized per 100 PGK1 copies, *p < 0.05 surface vs. crypt-base. n = 3 replicates). (G) Sum total of claudin transcript copies listed in D-F for each compartment (+p < 0.1, n = 3).
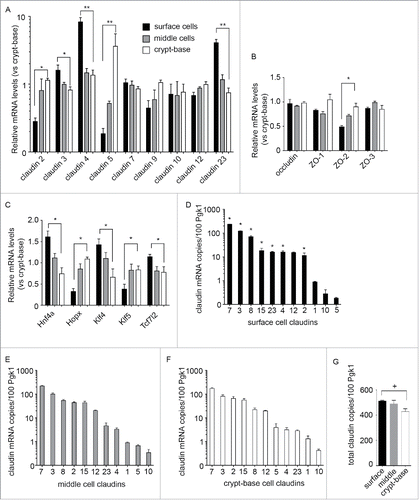
The above findings () are reported as fold change normalized to values in the crypt-base region. Epithelial barrier properties are dependent on the complement and relative proportion of claudin proteins in the tight junction.Citation26,27 Furthermore, with 24 Claudin genes in the mouse genome, not all claudin genes were screened on the array. Therefore, we wished to determine the abundance of all Cldn transcripts present along the crypt-base-surface epithelial cell axis. We performed a comprehensive real-time PCR screen of Cldn1-24 gene expression along the colonic crypts-base to surface cell axis (Fig. S1). All 24 claudins were examined by real time PCR and 10 abundant claudins (normalized to phosphoglycerate kinase 1 (Pgk1), (selection threshold 0.035 Pgk1) were selected for further analysis by quantitative ddPCR (). Cldn1 was also selected for further analysis due to its reported expression in intestinal epithelial cells.Citation28 Cldn14 and 20 were also detected in the crypt-base region, yet marginally expressed in proportion to Pgk1 (Table S3). With these analyses, we identified Cldn3 and 7 as the predominant claudins expressed in all zones along the crypt-base-to-surface cell axis, with Cldn2 approaching parity with Cldn3 in the crypt-base region (). Consistent with our previous results, the pore-forming Cldn2 was more abundant in the crypt-base, yet copy numbers were lower than those of Cldn7 (). Conversely, Cldn4 was found at high copy numbers in surface cell populations when compared to crypt-base (). Furthermore, quantitative PCR allowed for sum copy number comparisons for claudin family genes between zones along the crypt-base-to–surface axis (). Remarkably, total copy number of claudin family genes was found to fluctuate only slightly between compartments, with approximately 475 claudin transcript copies (per 100 Pgk1 copies, +/− 9%) expressed in each zone. This finding suggests that heterogeneous claudin gene family expression has a tightly regulated copy number along the crypt axis.
Hnf4a, Hopx, and Tcf7l2 regulate claudin expression in vitro
Since transcription factors orchestrate many cellular processes, we hypothesized that the TFs correlating with claudin expression are potential claudin regulators. In order to test this hypothesis, we overexpressed exogenous candidate TFs in model mouse intestinal epithelial CMT cells. Select claudins (as detected by qPCR/ddPCR in ) were then monitored for changes in expression by qPCR. CMT cells were transiently transfected with myc-tagged Hnf4a, Hopx, or Tcf7l2 and expression ensued for 24hrs. Western blot analysis of CMT cell lysates confirmed exogenous transcription factor expression (Fig. S2A/B). Additionally, TF nuclear localization was monitored by immunofluorescence (Fig. S2C). In parallel with these experiments, mRNA was harvested, allowing for the assessment of Cldn and TF gene expression in CMT cells. Throughout , a 1.5 threshold for biologically significant changes was applied, and illustrated by a dashed line.Citation29 In myc-Hnf4a overexpressing cells TF mRNA levels did not appreciably vary, and we observed low or non-detectable endogenous Hnf4a and Hopx (#, ). Myc-Hnf4a was only found to enhance Cldn23 gene expression, as shown in . Additionally, exogenous myc-Hopx expression was found to enhance Klf4, as well as several claudins (Cldn4, 7, 9 and 15), above the 1.5-fold change threshold (,D). However, high variability between replicates resulted in a failure to show statistical significance (p < 0.05). In myc-Tcf7l2 expressing cells, Hopx expression was also not detected (Ct > 33). However, we observed a marked increase in Hnf4a mRNA after Tcf7l2 transfection (). Enhanced Hnf4a was seen in all 3 replicate experiments, while endogenous Hnf4a signal was only detected in 1 of 3 experiments. While fold differences in expression could not be determined due to lack of endogenous detection, these data show that ectopic Tcf7l2 expression is sufficient to induce Hnf4a mRNA expression (). In claudin gene expression studies, myc-Tcf7l2 overexpression consistently and repeatedly stimulated Cldn2, 3, 4, 12, 15 and 23 (). In nine replicate experiments, we failed to detect Cldn1, 5, 8, or 10 in CMT cells (Ct > 33, ).
Figure 4. The transcription factors Hnf4a, Hopx, and Tcf7l2 regulate claudin message levels in vitro. (A-B) Myc-Hnf4a expression increases selectively Cldn23 relative to empty vector control (pCDNA). qPCR assessment of TF and claudin mRNA levels after Hnf4a expression in mouse intestinal cells (CMTs). Baseline Hnf4a mRNA was not detected in 1 out of 3 assays (#). Hopx expression was not detected (n.d.). Dashed line; 1.5-fold threshold (n = 3–4, *p < 0.05 by paired t-test, claudin 23 vs. pCDNA). (C-D) Myc-Hopx overexpression enhances Klf4 and Cldn4, 7 and 15. Dashed line; 1.5-fold threshold (n = 3). Baseline Hopx was not detected in 2 of 3 experiments (#). (E-F) Tcf7l2 regulates Cldn2, 3, 4, 12, 15 and 23. qPCR assessment of transcription factor and claudin gene expression in CMT cells following overexpression of myc-Tcf7l2. Baseline Hnf4a mRNA was not detected in 2 out of 3 assays (#). Dashed line; 1.5-fold threshold. Relative claudin mRNA expression in CMT cells after overexpression of myc-Tcf7l2 (n = 3–6. *p < 0.05 by paired t-test vs. pCDNA control). (G) Hnf4a is upregulated in myc-Tcf7l2 overexpressing cells (h2O, water only control).
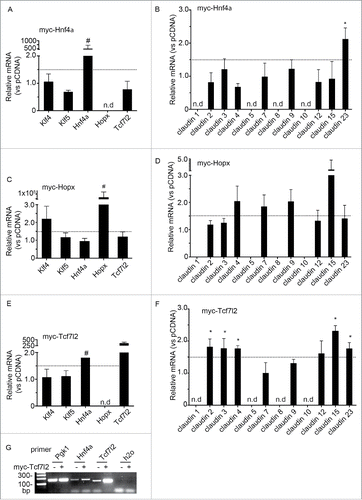
In summary, ectopic Hopx promotes Klf4 expression, while Tcf7l2 promotes Hnf4a. With respect to claudin regulation, Hnf4a overexpression increased the expression of only Cldn23. In contrast, CMT cells stimulated by myc-Hopx or myc-Tcf7l2 expression showed increased levels of several claudins. These findings demonstrate differential regulation of claudin genes by the TFs Hnf4a, Hopx, and Tcf7l2; Hnf4a appears to be a specific activator of Cldn23, with Hopx and Tcf7l2 having a more general claudin activating role.
Chromatin immunoprecipitation demonstrates Tcf7l2 binding at the Hnf4a promoter and Hnf4a binding at the Cldn23 promoter
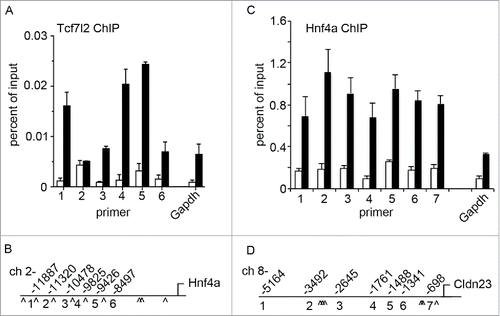
Our above findings demonstrate Tcf7l2 regulation of Hnf4a, as well as Hnf4a regulation of Cldn23. In order to determine if these factors play a direct role in gene regulation we analyzed Tcf7l2 and Hnf4a protein binding within promoter regions by Chromatin Immunoprecipitation (ChIP) (). A detailed in silico screen of the Hnf4a and Cldn23 promoters revealed several Tcf7l2 and Hnf4a consensus binding sites (). As shown in , Tcf7l2 antibodies co-precipitated with regions of the Hnf4a promoter. Additionally, Hnf4a ChIP co-precipitated Cldn23 promoter-specific DNA (). This was not the case for the non-specific rabbit IgG control ChIP, and limited binding was observed for genomic regions of the housekeeping gene Gapdh. These findings indicate that the transcription factors Tcf7l2 and Hnf4a directly bind to the Hnf4a and Cldn23 promoters respectively. These data show direct regulation of Hnf4a by Tcf7l2, and subsequent regulation of Cldn23 by Hnf4a.
Figure 5. Tcf7l2 and Hnf4a directly regulate Cldn23 expression. (A) Chromatin immunoprecipitation (ChIP) demonstrates Tcf7l2 association with the Hnf4a promoter. (B) Hnf4a promoter map indicating primer sites and approximate positions of putative Tcf7l2 binding sites (^). (C) Binding of Hnf4a to several sites in the Cldn23 promoter. (D) Cldn23 promoter map indicating primer sites and approximate positions of putative Hnf4a binding sites (^).
Hnf4a is required for differentiation dependent enhancement of Cldn23 in surface IECs
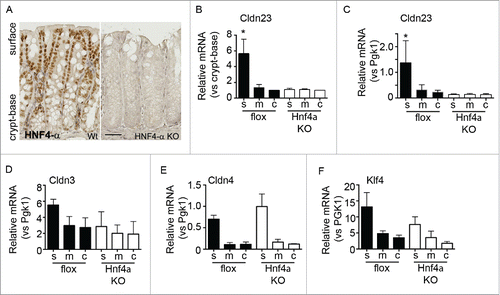
Hnf4a deficient mice have been reported to have defects in barrier function and claudin gene expression, however IEC claudin differentiation along the colonic crypt-to-surface epithelial cell axis was not assessed.Citation23 We therefore examined intestine specific Hnf4a conditional knockout mice (Hnf4alphaDeltaIEpC) for Cldn23 expression along the crypt-base–surface axis. As shown in , immunohistochemical staining of Hnf4a protein shows a gradient of nuclear staining intensity, with high levels in surface epithelia. High levels of Hnf4a are seen in the surface cell compartment relative to the crypt-base. Hnf4a knockout animals do not express functional protein and no signal could be detected by immunohistochemical staining (right panel, ). In order to determine the role of Hnf4a in Cldn23 regulation, serial cryosections were processed and analyzed for claudin gene expression. In keeping with our hypothesis, conditional Hnf4a knockout mice do not exhibit increased Cldn23 expression in surface cell populations as compared to crypt epithelial cells. This is in contrast to flox/flox littermate controls (). Indeed, when these values are normalized to the housekeeping gene Pgk1, we observe that Hnf4a deficient mice fail to induce expression of Cldn23 in surface IECs, in addition to overall lowered Cldn23 expression (). Along with a loss of Cldn23 induction, we observed aberrant Cldn3 and Klf4 expression in conditional Hnf4a knockout mice (). Conversely, Cldn4 levels were similar in the absence of Hnf4a, indicating claudin gene specific regulation by Hnf4a (). Importantly, Tcf7l2 transcript levels and protein localization were similar in Hnf4a knockout mice when compared with control animals (Fig. S3).
Figure 6. Hnf4a is required for surface expression of Cldn23. (A) Immunohistochemical staining of colonic tissue from flox/flox and intestinal specific Hnf4a knockout mice. Hnf4a protein is expressed in a gradient with higher expression in the surface cells. (B) qPCR of Cldn23 expression in surface(s), mid (m), and crypt-base (c) cell populations (normalized to crypt-base, n = 2, *p < 0.05 by 2-way ANOVA). (C) Analysis of Cldn23 expression relative to the housekeeping gene, Pgk1. (D-F) qPCR assessment of Cldn3, 4, and Klf4 in Hnf4a knockout mice.
Discussion
The complement of claudin protein expression plays an important role in controlling epithelial barrier properties and tissue homeostasis. Indeed, variations in claudin gene expression determine the physiological character of the paracellular TJ seal, and therefore the barrier function of the tissue. We now report a comprehensive account of claudin gene expression in mouse colonic mucosa during epithelial cell differentiation as they emerge from the crypt stem cell niche. Secondly, we describe 2 novel pathways that contribute to dynamic claudin expression throughout this process; Hopx and Tcf7l2/Hnf4a.
In order to discover novel regulators of claudin gene expression in colonic crypts, whole transcriptome analysis was performed on cells harvested from crypt-base and surface epithelial cell populations. This approach allowed us to identify the expression patterns of a number of claudin genes, as well as allowing for the identification of correspondingly expressed transcription factors. Our study also generated a wealth of information concerning the gene expression profiles in these spatially distinct compartments of the colon. Gene ontology analysis identified a number of pathways that contain these differentially expressed genes, the most significant of which are listed in Table S2. The predominant pathways identified involve cell cycle mediated processes, with the second most significant being cell adhesion. Interestingly, all the differentially expressed transcription factors that we identify as claudin regulators also play a role in regulating cell cycle. For example, Tcf7l2 is a Wnt signaling pathway effector and regulates glucose homeostasis in the gut.Citation30 Tcf7l2−/− mice die soon after birth and lack a proliferative stem cell compartment in the gut.Citation31 Several studies have identified Hnf4a as a regulator of intestinal viability and function.Citation32,33 For example, Hnf4a is protective in experimental colitis models and regulates colonic claudin 15-dependent ion transport.Citation23 Additionally, Hnf4a defective mice develop spontaneous adenomas.Citation34,35 Although Klf4−/− mice exhibited normal cell proliferation in the gut, there is evidence that Klf4 is an inhibitor of β-catenin signaling.Citation36,37 Hopx has been reported to play a role in colorectal cancer, and its expression is sustained in colon cancers.Citation38 Furthermore, Hopx is a stem cell marker within the 4+ cell region transient amplifying zone.Citation39 Together with our findings, these studies demonstrate intriguing co-regulation of proliferative processes and claudin gene expression in the colonic epithelium.
Defects in the claudin-based TJ barrier are believed to contribute to the epithelial barrier compromise in inflammatory bowel diseases by allowing mucosal exposure to luminal antigens.Citation40 Additionally, failure to regulate claudin gene expression also contributes to carcinogenesis, both directly and/or secondarily, by altering the cellular microenvironment.Citation41 Further roles for claudins have also been described implicating them in the pathogenesis of cancer, as dysregulation of claudin expression has also been linked to epithelial to mesenchymal transition (EMT). Interestingly, claudins linked to dysregulated EMT include Claudin 4 and 7,Citation42-44 which we find to be enriched in differentiated IECs. These claudins are found at lower levels in cancer cells, indicating either a direct role for these claudins in regulating these processes or signify a state of poor epithelial differentiation.Citation42 Further studies will be required to understand the role of claudin proteins in health and disease.
Our study provides the first comprehensive description of claudin gene expression along the colonic crypt-base-to-surface axis in mice and serves as a reference for further studies of genes involved in the epithelial differentiation process. Surface epithelial cell populations are enriched in cells expressing Cldn3, 4, 7 and 23, while crypt-base cells express Cldn2, 5, and 15 (). Cldn 1, 10 and 12 appear to be expressed ubiquitously along the crypts. Cldn14 and 20 were also crypt enriched, yet found to be marginally expressed relative to other claudins (Fig. S1). It is interesting to note that the claudin genes identified as differentially expressed fall into one of 2 functional groups, “leaky” pore forming claudins in the crypt-base (Cldn2, 5, 14, and 15) and “tight” sealing claudins in the surface cell populations (Cldn3 and 7).Citation11,45 Cldn23 was observed to be enriched in surface IECs suggesting a role in barrier tightening.
Figure 7. Summary of claudins detected and model of signaling pathway for Cldn23 expression. (A) Claudin expression gradients along the crypt to surface axis. Surface enriched claudins in blue. Crypt enriched claudins in red (* identified by qPCR only). (B) Schematic demonstrating the relative abundance of Cldn genes along the crypt-base to surface axis. (C) Schematic showing Hopx/Klf4 regulation of Cldn4 and 7 as well as Tcf7l2 stimulation of Hnf4a which in turn activates Cldn23.
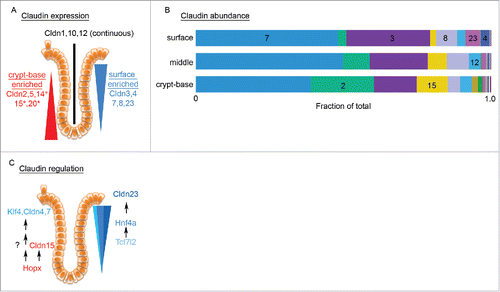
In regard to relative transcript proportions, Cldn7 predominates throughout the entire colonic crypt, followed by Cldn3, with Cldn2 reaching parity with Cldn3 in the crypt-base region (). Interestingly, we found very little difference at the copy number level, between regions of the colonic crypt. This finding supports the hypothesis that heterogeneous claudin gene family expression has a tightly regulated copy number limit, with specific regulators within each compartment determining which claudin transcripts are expressed. Based on our studies, we propose that Hopx and Tcf7l2 are general activators of claudin transcription due to the lack of specific spatiotemporal claudin gene expression. This is demonstrated by overexpression experiments in which the crypt-base specific Cldn2 was stimulated in addition to surface specific claudins such as Cldn3, 4 and 7 (). Similarly, Hopx stimulated both the crypt restricted Cldn15 as well as the surface enriched Cldn4 and 9. In contrast, in CMT cells, Hnf4a was found to selectively upregulate Cldn23.
Our array data indicate that Tcf7l2 is enriched in crypt-base populations, however, our qPCR findings show the opposite trend, with higher amounts of Tcf7l2 in surface populations (). This discrepancy may be the result of differences in Tcf7l2 isoforms expressed in the 2 compartments. Indeed, Tcf7l2 is known to undergo extensive splicing.Citation46 However, our studies and others have shown Tcf7l2 protein accumulating in surface epithelial cell populations.Citation37 Further discrepancies arose between the array and qPCR methodologies; the ZO and occludin TJ proteins could not be confirmed as expressed in a gradient. These exceptions notwithstanding, we observed a high degree of similarity between our array and qPCR findings.
Tcf7l2 was found to activate the transcription of Hnf4a in CMT cells (). As discussed above, endogenous Hnf4a in this cell line could not be detected in the majority of samples tested, indicating that this factor is at low endogenous levels. Importantly, Hnf4a overexpression in CMT cells was sufficient to stimulate mRNA expression of Cldn23 (). Claudin 23 is a poorly studied claudin but has been found to be suppressed in gastric cancers as well as colon cancer.Citation47,48 We now report that Cldn23 is abundant in its expression in colonic IEC surface epithelial cell populations, and using ChIP, have identified Hnf4a as a binding factor of the Cldn23 promoter. In conclusion, our data support a claudin differentiation program that includes Tcf7l2 stimulation of Hnf4a, and the subsequent activity of both agents to stimulate claudin gene expression, including Cldn23 ().
Materials and methods
Mice
All experimental protocols used male, 10–12 week old, C57BL/6 (WT) mice (The Jackson Laboratory, Bar Harbor, Me) or intestinal epithelial cell specific conditional Hnf4a KO mice (Hnf4alpha(DeltaIEpC), from Dr F.J. Gonzalez ) as describedCitation35,49 and littermate flox/flox (flanked loxP) controls. All procedures using animals were reviewed and approved by the Emory University Institutional Animal Care and Use Committee and were performed according to National Institutes of Health criteria.
Tissue collection by LCM, RNA extraction, and amplification
Distal colon rolls were prepared, cryosectioned, and fixed using the Arcturus Histogene kit for frozen sections (Applied Biosystems/Life Technologies, Grand Island, NY). The Arcturus Laser Capture Microdissection (LCM) system (Applied Biosystems/Life Technologies) was used to isolate crypt-base and surface cell populations. Sequential isolation of crypt-base and surface cell populations occurred for each of the 4 mice. RNA extraction was then performed according to the manufacturer's instruction using the Arcturus PicoPure RNA extraction kit (Applied Biosystems/Life Technologies), treated with DNase (Qiagen, Valencia, CA), and amplified twice using a Nugen WT-Ovation kit (San Carlos, CA). RNA integrity was confirmed by Agilent 2100 Bioanayzer (Santa Clara, CA).
Cell/Tissue collection and RNA extraction for real-time PCR validation
Confirmation of microarray candidates was performed on RNA extracted from serial cryosections of whole mount distal colonic tissue as previously described.Citation21 Briefly, distal colon samples were obtained from a 2 cm segment spanning 2–4 cm from the anus. RNA was extracted using Trizol reagent according to manufacturer's instruction (Life Technologies), subjected to secondary purification (Epoch Life Sciences, Sugarland TX), DNAse treatment (Qiagen), and converted to cDNA (Thermo Fisher, Waltham, MA). Semi-quantitative real time PCR was performed with iQ Sybr Green (BioRad,) using a BioRad iCycler (BioRad). Quantitative PCR was performed on a QX200 ddPCR system (BioRad). Primers used are listed in Table S1. Primers were validated by product sequencing and efficiency test (Efficiency=1.00±20%), unless validated elsewhere. QPCR data processing utilized a delta delta Cq method, with a 1.5 fold mean threshold for biological significant change. This value exceeds threshold assessment performed in Table S3, as described previously.Citation29
Microarray data analysis
Eight samples (4 replicates from the crypt-base and 4 replicates from the surface) from the Illumina MouseWG-6_v 1.1 platform were processed using the default Illumina Genome Studio software for normalization and summarization. All probes with a detection p-value < 0.05 in each of the 8 samples were considered detected. The expression values from all samples were log2 transformed and then averaged for each probe across each sample type, so that for each probe there was an average crypt-base and an average crypt-surface value. The fold change was then calculated by subtracting the average value of the crypt-base replicates minus the average value of the crypt-surface replicates. A positive fold change indicated upregulation within the crypt-base and conversely, a negative fold change indicated down-regulation at the crypt-base compared to the surface. From the initial 46,643 probes, 35,371 had nearly constant expression values (standard deviation ≤ 0.2) across all 8 samples and were excluded from the differential expression analysis. The remaining 11,272 probes were used to calculate the differentially expressed genes. The two-sample, 2-tail, unequal variance t-test was used for the statistical comparison and the standard p-value cutoff gave 2,731 differentially expressed genes. The p-values were further corrected for multiple comparisons with the FDR (false discovery) method. The FDR cutoff value of 0.2 resulted in 2,639 statistically significant differentially expressed genes between the crypt-base and the crypt-surface compartments. These genes were used for downstream functional analyses. For pathway analysis, the GeneGO software was utilized (https://portal.genego.com/). For the hierarchical clustering and the generation of the heatmaps, the R packages “dplyr” and “NMF” were downloaded from the CRAN site (https://cran.r-project.org/web/packages/). The Euclidean distance and the average agglomeration method were used for clustering the genes and samples. For the transcription factor identification, data from the RIKEN database (http://genome.gsc.riken.jp/TFdb/) and the Animal Transcription Factor Database (http://www.bioguo.org/AnimalTFDB/) were combined. The Pearson's correlation coefficient along with the associated p-values were calculated using the R function cor.test. Microarray data submission to NCBI GEO (GEO number GSE84742) is in progress.
Chromatin immunoprecipitation
Chromatin Immunoprecipitation (ChIP) was performed using the Magna-ChIP kit (EMD Millipore, Billerica, MA, USA) according to the manufacturer's recommendations. Confluent monolayers of CMT cells were fixed with fresh 1 % formaldehyde solution, quenched with glycine and collected in ice cold PBS containing protease inhibitors. The cells were then lysed in ChIP lysis buffer and the chromatin was sheared with a Branson 450 Sonifier (Branson Ultrasonics, Danbury CT, USA) on ice. The sonicator power was set to 3 and 30 % duty cycle. Sonication was performed 4 times for 30 seconds with a one minute breaks to prevent heat accumulation. This resulted in uniform chromatin shearing to approximately 200–500 bp. Twenty-five µg chromatin was used per IP using 300 ng TCF7l2 or 5 μg anti-HNF4a (Cell Signaling, Boston, MA) in parallel to nonspecific anti rabbit IgG. After reversing the crosslinking and digestion with proteinase-K, DNA was isolated using spin columns. qPCR primers were designed to amplify DNA sequences spanning putative TF binding. Putative TF binding sites were identified using PROMO (http://alggen.lsi.upc.es/cgi-bin/promo_v3/promo/promoinit.cgi?dirDB=TF_8.3).
Tissue culture and constructs
CMT (CMT-93, ATCC, Mannassas VA) and SKCO15 Citation50 cells were cultured in DMEM media with 10% FBS and antibiotics (cells were periodically tested for mycoplasma infection). Murine Hopx was cloned from C57/BL6 male mouse colonic cDNA: (Forward: primer aagcttgccaccatgtcggcgcagaccgcgagcg and Reverse primer: ggatccgtccgtaacagatctgcattccg) and ligated into pcDNA3.1. All constructs were sequenced to confirm proper construction of the plasmids. Plasmids were generated by the Emory Cloning Core.
Antibodies and reagents
Western Blot and immunofluorescence (IF) analysis antibodies include: as a protein loading control glyceraldehyde-3-phosphate dehydrogenase (GAPDH1(G8795), 1:5000), 9E10 anti-myc,Citation1 1:5000, TCF4Citation3 (C48H11), 1:500, Hnf4aCitation3 (C11F12), 1:500, and HRP/fluorophore-linked secondary antibodies,Citation4 1:10,000 (1 Sigma Aldrich, St. Louis, MO;2 Invitrogen, Carlsbad, CA;3 Cell Signaling, Beverly, MA;4 Jackson ImmunoResearch, Westgrove).
Immunofluorescence (IF) staining and microscopy
Cells were fixed in 3.7% paraformaldehyde or 100% methanol for 20 minutes. Primary antibody reactions were performed in HBSS+/+ with 3% BSA for 1hr. Secondary antibodies (Alexa Fluor 546-conjugated) were incubated 3% BSA and for 45 min. Nuclei were detected with TOPRO-3. Confocal microscopy was performed using a Zeiss LSM 510 microscope (Zeiss, Thornwood, NY).
Western blot analysis
Cells were lysed in lysis buffer (50 mM Tris (pH 6.8), 10% glycerol, 2% SDS) supplemented with phosphatase inhibitor cocktail I and II, and protease inhibitor cocktail (Sigma Aldrich, St. Louis, MO), incubated at 60C for 30min, and subjected to SDS-PAGE. Primary antibodies were incubated for 1hr at RT followed by a 3x wash with TTBS. HRP-conjugated secondary antibodies were incubated at RT for 45 minutes.
Statistics
Statistical analysis for qPCR/ddPCR and TEER data were performed as indicated using GraphPad Prism (GraphPad Software Inc., La Jolla CA). Analysis of variance (ANOVA) includes Bonferroni post-test. Student t-test or Wilcoxon-Mann-Whitney test as indicated. Error is reported as standard error of the mean (SEM).
Summary statement
Exposure to the intestinal milieu contributes to a number of pathologies. The claudin gene family forms a protective barrier through coordinated expression via Hopx, Tcf7l2 and Hnf4a.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Author contributions
L.L. and A.F. performed the experiments with assistance from C.G-S. C.E.O. performed experiments in . V.H. and R.H. . C.M. reviewed microarray data. C.T.C. designed the study, performed experiments, wrote the main manuscript text, and prepared the figures, with input from C.A.P. and A.N. All authors reviewed the manuscript.
KTIB_S_1214038.zip
Download Zip (4.4 MB)Acknowledgments
We would like to thank Barbara Hrdlickova for discussion and review of this manuscript. We would also like to thank Oskar Laur at the Emory University Cloning Core and Dr. F.J. Gonzalez for providing the Hnf4alphaDeltaIEpC mice.
Funding
Our studies were supported by National Institutes of Health (DK55679 and DK59888 to A.N., DK061379 and DK072564 to CAP), and the Crohn's and Colitis Foundation of America (CCFA) Career Development Award to C.T.C. and CCFA Fellowship Award to A.E.F.
References
- Van Itallie CM, Anderson JM. Architecture of tight junctions and principles of molecular composition. Semin Cell Dev Biol 2014; 36C:157-165; ; http://dx.doi.org/10.1016/j.semcdb.2014.08.011
- Colegio OR, Van Itallie CM, McCrea HJ, Rahner C, Anderson JM. Claudins create charge-selective channels in the paracellular pathway between epithelial cells. Am J Physiol Cell Physiol 2002; 283:C142-147; PMID:12055082; http://dx.doi.org/10.1152/ajpcell.00038.2002
- Tsukita S, Furuse M. Pores in the wall: claudins constitute tight junction strands containing aqueous pores. J Cell Biol 2000; 149:13-16; PMID:10747082; http://dx.doi.org/10.1083/jcb.149.1.13
- Humphries A, Wright NA. Colonic crypt organization and tumorigenesis. Nat Rev Cancer 2008; 8:415-424; PMID:18480839; http://dx.doi.org/10.1038/nrc2392
- A3B2 twb=.2w?>Weber CR, Turner JR. Inflammatory bowel disease: is it really just another break in the wall? Gut 2007; 56:6-8; PMID:17172583; http://dx.doi.org/10.1136/gut.2006.104182
- Schlingmann B, Molina SA, Koval M. Claudins: Gatekeepers of lung epithelial function. Semin Cell Dev Biol 2015; 42:47-57; PMID:25951797; http://dx.doi.org/10.1016/j.semcdb.2015.04.009
- Krug SM, Schulzke JD, Fromm M. Tight junction, selective permeability, and related diseases. Sem Cell Dev Biol 2014; 36C:166-176; ; http://dx.doi.org/10.1016/j.semcdb.2014.09.002
- Yu AS. Claudins and the kidney. J Am Soc Nephrol 2015; 26:11-19; PMID:24948743; http://dx.doi.org/10.1681/ASN.2014030284
- Wada M, Tamura A, Takahashi N, Tsukita S. Loss of claudins 2 and 15 from mice causes defects in paracellular Na+ flow and nutrient transport in gut and leads to death from malnutrition. Gastroenterology 2013; 144:369-380; PMID:23089202; http://dx.doi.org/10.1053/j.gastro.2012.10.035
- Lu Z, Ding L, Lu Q, Chen YH. Claudins in intestines: Distribution and functional significance in health and diseases. Tissue Barr 2013; 1:e24978; ; http://dx.doi.org/10.4161/tisb.24978
- Capaldo CT, Nusrat A. Claudin switching: Physiological plasticity of the Tight Junction. Semin Cell Dev Biol 2015; 42:22-9; PMID:25957515; http://dx.doi.org/10.1016/j.semcdb.2015.04.003
- Noah TK, Donahue B, Shroyer NF. Intestinal development and differentiation. Exp Cell Res 2011; 317:2702-2710; PMID:21978911; http://dx.doi.org/10.1016/j.yexcr.2011.09.006
- Barker N. Adult intestinal stem cells: critical drivers of epithelial homeostasis and regeneration. Nat Rev Mol Cell Biol 2014; 15:19-33; PMID:24326621
- Stappenbeck TS, Mills JC, Gordon JI. Molecular features of adult mouse small intestinal epithelial progenitors. Proc Natl Acad Sci U S A 2003; 100:1004-1009; PMID:12552106; http://dx.doi.org/10.1073/pnas.242735899
- Mariadason JM, Arango D, Shi Q, Wilson AJ, Corner GA, Nicholas C, Aranes MJ, Lesser M, Schwartz EL, Augenlicht LH. Gene expression profiling-based prediction of response of colon carcinoma cells to 5-fluorouracil and camptothecin. Cancer Res 2003; 63:8791-8812; PMID:14695196
- Giannakis M, Stappenbeck TS, Mills JC, Leip DG, Lovett M, Clifton SW, Ippolito JE, Glasscock JI, Arumugam M, Brent MR, et al. Molecular properties of adult mouse gastric and intestinal epithelial progenitors in their niches. J Biol Chem 2006; 281:11292-11300; PMID:16464855; http://dx.doi.org/10.1074/jbc.M512118200
- Kosinski C, Li VS, Chan AS, Zhang J, Ho C, Tsui WY, Chan TL, Mifflin RC, Powell DW, Yuen ST, et al. Gene expression patterns of human colon tops and basal crypts and BMP antagonists as intestinal stem cell niche factors. Proc Natl Acad Sci U S A 2007; 104:15418-15423; PMID:17881565; http://dx.doi.org/10.1073/pnas.0707210104
- Darido C, Buchert M, Pannequin J, Bastide P, Zalzali H, Mantamadiotis T, Bourgaux JF, Garambois V, Jay P, Blache P, et al. Defective claudin-7 regulation by Tcf-4 and Sox-9 disrupts the polarity and increases the tumorigenicity of colorectal cancer cells. Cancer Res 2008; 68:4258-4268; PMID:18519685; http://dx.doi.org/10.1158/0008-5472.CAN-07-5805
- Escaffit F, Boudreau F, Beaulieu JF. Differential expression of claudin-2 along the human intestine: Implication of GATA-4 in the maintenance of claudin-2 in differentiating cells. J Cell Physiol 2005; 203:15-26; PMID:15389642; http://dx.doi.org/10.1002/jcp.20189
- Bhat AA, Sharma A, Pope J, Krishnan M, Washington MK, Singh AB, Dhawan P. Caudal homeobox protein Cdx-2 cooperates with Wnt pathway to regulate claudin-1 expression in colon cancer cells. PloS one 2012; 7:e37174; PMID:22719836; http://dx.doi.org/10.1371/journal.pone.0037174
- Farkas AE, Gerner-Smidt C, Lili L, Nusrat A, Capaldo CT. Cryosectioning method for microdissection of murine colonic mucosa. J Visual Exp 2015; 101:e53112; PMID:26274554; http://dx.doi.org/10.3791%2F53112
- Farkas AE, Hilgarth RS, Capaldo CT, Gerner-Smidt C, Powell DR, Vertino PM, Koval M, Parkos CA, Nusrat A. HNF4alpha regulates claudin-7 protein expression during intestinal epithelial differentiation. Am J Pathol 2015; 185:2206-2218; PMID:26216285; http://dx.doi.org/10.1016/j.ajpath.2015.04.023
- Darsigny M, Babeu JP, Dupuis AA, Furth EE, Seidman EG, Lévy E, Verdu EF, Gendron FP, Boudreau F. Loss of hepatocyte-nuclear-factor-4alpha affects colonic ion transport and causes chronic inflammation resembling inflammatory bowel disease in mice. PloS one 2009; 4:e7609; PMID:19898610; http://dx.doi.org/10.1371/journal.pone.0007609
- Ghaleb AM, Laroui H, Merlin D, Yang VW. Genetic deletion of Klf4 in the mouse intestinal epithelium ameliorates dextran sodium sulfate-induced colitis by modulating the NF-kappaB pathway inflammatory response. Inflammat Bowel Dis 2014; 20:811-820; ; http://dx.doi.org/10.1097/MIB.0000000000000022
- McConnell BB, Kim SS, Bialkowska AB, Yu K, Sitaraman SV, Yang VW. Kruppel-like factor 5 protects against dextran sulfate sodium-induced colonic injury in mice by promoting epithelial repair. Gastroenterology 2011; 140:540-549 e542; PMID:21078320; http://dx.doi.org/10.1053/j.gastro.2010.10.061
- Koval M. Differential pathways of claudin oligomerization and integration into tight junctions. Tissue Barr 2013; 1:e24518; PMID:24665398; http://dx.doi.org/10.4161/tisb.24518
- Furuse M, Furuse K, Sasaki H, Tsukita S. Conversion of zonulae occludentes from tight to leaky strand type by introducing claudin-2 into Madin-Darby canine kidney I cells. J Cell Biol 2001; 153:263-272; PMID:11309408; http://dx.doi.org/10.1083/jcb.153.2.263
- Pope JL, Bhat AA, Sharma A, Ahmad R, Krishnan M, Washington MK, Beauchamp RD, Singh AB, Dhawan P. Claudin-1 regulates intestinal epithelial homeostasis through the modulation of Notch-signalling. Gut 2014; 63:622-634; PMID:23766441; http://dx.doi.org/10.1136/gutjnl-2012-304241
- Edmunds RC, McIntyre JK, Luckenbach JA, Baldwin DH, Incardona JP. Toward enhanced MIQE compliance: reference residual normalization of qPCR gene expression data. J Biomol Tech 2014; 25:54-60; PMID:24982597
- Shao W, Wang D, Chiang YT, Ip W, Zhu L, Xu F, Columbus J, Belsham DD, Irwin DM, Zhang H, et al. The Wnt signaling pathway effector TCF7L2 controls gut and brain proglucagon gene expression and glucose homeostasis. Diabetes 2013; 62:789-800; PMID:22966074; http://dx.doi.org/10.2337/db12-0365
- Korinek V, Barker N, Moerer P, van Donselaar E, Huls G, Peters PJ, Clevers H. Depletion of epithelial stem-cell compartments in the small intestine of mice lacking Tcf-4. Nat Genet 1998; 19:379-383; PMID:9697701; http://dx.doi.org/10.1038/1270
- Cattin AL, Le Beyec J, Barreau F, Saint-Just S, Houllier A, Gonzalez FJ, Robine S, Pinçon-Raymond M, Cardot P, Lacasa M, et al. Hepatocyte nuclear factor 4alpha, a key factor for homeostasis, cell architecture, and barrier function of the adult intestinal epithelium. Mol Cell Biol 2009; 29:6294-6308; PMID:19805521; http://dx.doi.org/10.1128/MCB.00939-09
- San Roman AK, Aronson BE, Krasinski SD, Shivdasani RA, Verzi MP. Transcription factors GATA4 and HNF4A control distinct aspects of intestinal homeostasis in conjunction with transcription factor CDX2. J Biol Chem 2015; 290:1850-1860; PMID:25488664; http://dx.doi.org/10.1074/jbc.M114.620211
- Chahar S, Gandhi V, Yu S, Desai K, Cowper-Sal-lari R, Kim Y, Perekatt AO, Kumar N, Thackray JK, Musolf A, et al. Chromatin profiling reveals regulatory network shifts and a protective role for hepatocyte nuclear factor 4alpha during colitis. Mol Cell Biol 2014; 34:3291-3304; PMID:24980432; http://dx.doi.org/10.1128/MCB.00349-14
- Ahn SH, Shah YM, Inoue J, Morimura K, Kim I, Yim S, Lambert G, Kurotani R, Nagashima K, Gonzalez FJ, et al. Hepatocyte nuclear factor 4alpha in the intestinal epithelial cells protects against inflammatory bowel disease. Inflammat Bowel Dis 2008; 14:908-920; ; http://dx.doi.org/10.1002/ibd.20413
- Zhang W, Chen X, Kato Y, Evans PM, Yuan S, Yang J, Rychahou PG, Yang VW, He X, Evers BM, et al. Novel cross talk of Kruppel-like factor 4 and beta-catenin regulates normal intestinal homeostasis and tumor repression. Mol Cell Biol 2006; 26:2055-2064; PMID:16507986; http://dx.doi.org/10.1128/MCB.26.6.2055-2064.2006
- Evans PM, Chen X, Zhang W, Liu C. KLF4 interacts with beta-catenin/TCF4 and blocks p300/CBP recruitment by beta-catenin. Mol Cell Biol 2010; 30:372-381; PMID:19901072; http://dx.doi.org/10.1128/MCB.00063-09
- Yamashita K, Katoh H, Watanabe M. The homeobox only protein homeobox (HOPX) and colorectal cancer. Int J Mol Sci 2013; 14:23231-23243; PMID:24287901; http://dx.doi.org/10.3390/ijms141223231
- Takeda N, Jain R, LeBoeuf MR, Wang Q, Lu MM, Epstein JA. Interconversion between intestinal stem cell populations in distinct niches. Science 2011; 334:1420-1424; PMID:22075725; http://dx.doi.org/10.1126/science.1213214
- Schulzke JD, Ploeger S, Amasheh M, Fromm A, Zeissig S, Troeger H, Richter J, Bojarski C, Schumann M, Fromm M. Epithelial tight junctions in intestinal inflammation. Annal New York Acad Sci 2009; 1165:294-300; ; http://dx.doi.org/10.1111/j.1749-6632.2009.04062.x
- Ding L, Lu Z, Lu Q, Chen YH. The claudin family of proteins in human malignancy: a clinical perspective. Cancer Manag Res 2013; 5:367-375; PMID:24232410
- Kwon MJ. Emerging roles of claudins in human cancer. Int J Mol Sci 2013; 14:18148-18180; PMID:24009024; http://dx.doi.org/10.3390/ijms140918148
- Pope JL, Ahmad R, Bhat AA, Washington MK, Singh AB, Dhawan P. Claudin-1 overexpression in intestinal epithelial cells enhances susceptibility to adenamatous polyposis coli-mediated colon tumorigenesis. Mol Cancer 2014; 13:167; PMID:24997475; http://dx.doi.org/10.1186/1476-4598-13-167
- Bhat AA, et al. Claudin-7 expression induces mesenchymal to epithelial transformation (MET) to inhibit colon tumorigenesis. Oncogene 2015;34(35):4570-80; PMID:25500541; http://dx.doi.org/10.1038/onc.2014.385
- Baker M, Reynolds LE, Robinson SD, Lees DM, Parsons M, Elia G, Hodivala-Dilke K. Stromal Claudin14-heterozygosity, but not deletion, increases tumour blood leakage without affecting tumour growth. PloS one 2013; 8:e62516; PMID:23675413; http://dx.doi.org/10.1371/journal.pone.0062516
- Weise A, Bruser K, Elfert S, Wallmen B, Wittel Y, Wöhrle S, Hecht A. Alternative splicing of Tcf7l2 transcripts generates protein variants with differential promoter-binding and transcriptional activation properties at Wnt/beta-catenin targets. Nucl Acids Res 2010; 38:1964-1981; PMID:20044351; http://dx.doi.org/10.1093/nar/gkp1197
- Katoh M, Katoh M. CLDN23 gene, frequently down-regulated in intestinal-type gastric cancer, is a novel member of CLAUDIN gene family. Int J Mol Med 2003; 11:683-689; PMID:12736707
- Maryan N, Statkiewicz M, Mikula M, Goryca K, Paziewska A, Strzałkowska A, Dabrowska M, Bujko M, Ostrowski J. Regulation of the expression of claudin 23 by the enhancer of zeste 2 polycomb group protein in colorectal cancer. Mol Med Rep 2015; 12:728-736; PMID:25695204
- Hayhurst GP, Lee YH, Lambert G, Ward JM, Gonzalez FJ. Hepatocyte nuclear factor 4alpha (nuclear receptor 2A1) is essential for maintenance of hepatic gene expression and lipid homeostasis. Mol Cell Biol 2001; 21:1393-1403; PMID:11158324; http://dx.doi.org/10.1128/MCB.21.4.1393-1403.2001
- Yoo BK, Yanda MK, No YR, Yun CC. Human intestinal epithelial cell line SK-CO15 is a new model system to study Na(+)/H(+) exchanger 3. Am J Physiol Gastrointest liver Physiol 2012; 303:G180-188; PMID:22556145; http://dx.doi.org/10.1152/ajpgi.00069.2012
- Capaldo CT, Farkas AE, Hilgarth RS, Krug SM, Wolf MF, Benedik JK, Fromm M, Koval M, Parkos C, Nusrat A. Proinflammatory cytokine-induced tight junction remodeling through dynamic self-assembly of claudins. Mol Biol Cell 2014; 25:2710-2719; PMID:25031428; http://dx.doi.org/10.1091/mbc.E14-02-0773
