Abstract
Circular RNAs (circRNAs) have been revealed to play vital roles in modulating gene expression and participate in several pathological responses including gastric cancer (GC). However, the larger numbers of the circRNAs in GC progression remain undetermined. In the present study, four GC related circRNAs expression profiles from the Gene Expression Omnibus (GEO) database were enrolled. We identified hsa_ circRNA_00067997 (circ_0067997) as an oncogene in GC. qRT-PCR was used to validate the expression of circ_0067997 in GC tissues and cell lines. The results revealed that circ_0067997 was upregulated in GC, and high circ_0067997 expression was associated with the poor overall survival rate of GC patients. Knockdown of circ_0067997 significantly reduced cell viability, inhibited colony formation, and attenuated invasive ability, whereas overexpression of circ_0067997 exhibited opposed effects. Circ_0067997 was identified to be a sponge for miR-515-5p directly. Moreover, XIAP was demonstrated to be targeted and regulated by miR-515-5p. In conclusion, circ_0067997 was identified to be an oncogene in GC by regulating miR-515-5p/XIAP axis.
Introduction
Gastric cancer is considered to be the primary cause of tumour-related death worldwide [Citation1]. Although GC-related fatality rate was significantly downregulated these decades, the International Agency for Research on Cancer (IARC) data showed that GC remained to be the fifth most common malignancy [Citation2,Citation3]. It was estimated that around one million new cases were diagnosed as GC every year, and more than half of which was occurred in the Asia countries, such as China, Japan, and Korea [Citation1,Citation4]. Due to the limitation of current diagnostic techniques, a considerable amount of GC patients was finally confirmed at the advanced clinical stages, missing the best therapeutic opportunity [Citation5]. So, it is essential to elucidate the potential pathogenesis of GC, and explore several effective diagnostic biomarkers and therapeutic targets for GC patients.
Circular RNAs (circRNAs), formed by the head-to-tail splicing of several exons of target gene, is emerging as a new member of non-coding RNAs (ncRNAs) family [Citation6]. Due to its specific formation way, circRNAs usually present as covalently closed loop structures without 5′ caps and 3′ tail [Citation7]. It was well documented that microRNAs (miRNAs) and long non-coding RNAs (lncRNAs), which also belong to non-coding RNAs, involved in the tumorigenesis of multiple human tumours [Citation8,Citation9]. Recently, circRNAs were also revealed as oncogenes or tumour suppressors in various human cancers [Citation10–12]. Moreover, increased evidences revealed that circRNAs expression profiles were altered in GC by performing circRNAs microarray in GC tissues and corresponding normal tissues [Citation13]. For instance, results from a microarray carried out by Zou X et al. in three pairs of GC and normal samples suggested that a total of 347 increased and 603 decreased cricRNAs were dysregulated [Citation14]. Weng Y et al. reported that a total of 100 circRNAs were dysregulated in GC tissues compared to adjacent normal tissues [Citation15]. However, the detailed mechanism and functions of circRNAs in GC remain unclear.
In the present study, we enrolled four GSE datasets and identified that circ_0067997 was upregulated in GC samples. Then, we performed functional assays to explore its effects on GC cell viability, proliferation and invasion. In mechanisms, we predicted the target miRNAs of circ_0067997 and found miR-515-5p could be regulated by this circRNA. Finally, we revealed the downstream effector of the circ_0067997/miR-515-5p axis.
Material and methods
Patient samples, cell lines and transfection
A total of 48 pairs GC and corresponding non-cancer tissues were collected at The People's Hospital of Anyang city from 2014 to 2018 and maintained at −80 °C until further study. Manipulates of this study were approved by the ethics committee of the People’s Hospital of Anyang city, and written consents were obtained from all subjects. Normal human gastric epithelial mucosa cell line (GSE-1) and four human GC cell lines (SGC-7901, MGC-803, BGC-823 and MKN28) were all provided by the Type Culture Collection of Chinese Academy of Sciences (Shanghai, China) and were maintained following the instructions provided by providers.
To block the expression of circ_0067997 and miR-515-5p in GC cells, two siRNAs target circ_0067997 (si-circ_0067997-1 and si-circ_0067997-2) and miR-515-5p inhibitor were utilized. GC cells (1 × 105 cells/well) were seeded into 6-well plates until 80% confluent, si-circ_0067997-1/2 and miR-515-5p inhibitor were transfected into GC cells by Lipofectamine 3000 (Invitrogen, USA) following the manufacturer’s protocols. To overexpress the expression of circ_0067997 and XIAP in GC cells, the sequence of circ_0067997 and XIAP were synthesized and sub-cloned into pcDNA3.0 vector and then packed into lentivirus, which were then used to transfect GC cells. MiR-515-5p mimics were utilized to overexpress miR-515-5p in GC cell using Lipofectamine 3000 (Invitrogen, Waltham, MA, USA).
Differential expression analysis
Four gene expression profiles of GC (GSE78092, GSE83521, GSE89143 and GSE93541) were downloaded from the Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo). Online tool, GEO2R was used to analyze differentially expressed circRNAs between GC and normal samples in these GSE datasets. The adjusted p < .01 and FC >2 were set as the cut off criterion.
Pathway enrichment analysis
The target genes of miR-127–5, miR-451, miR-515-5p, miR-615-5p and miR-625 were analyzed by Kyoto Encyclopedia of Genes and Genomes (KEGG) via the Database for Annotation, Visualization and Integrated Discovery (DAVID, https://david.ncifcrf.gov/).
RNA extraction and quantitative real-time PCR (qRT-PCR) assay
Total RNAs of GC and corresponding non-cancer tissues, as well as treated GC cell lines, were extracted using TRIzol reagent (#9109, Takara, Japan). Reverse transcription was carried out via BestarTM qPCR RT kit (#2220, DBI Bioscience, China) based on 5 μg of total RNAs. The real-time quantitative PCR amplification was performed on ABI7000 system by BestarTM qPCR MasterMix (#2043, DBI Bioscience, China) following the protocols provided by the manufacturer. The sequence of primers used in the present study is shown in . The expression of miRNAs was normalized to U6, and the mRNA expression of cicr_0067997 and XIAP were normalized to GAPDH.
TABLE 1. Primer sequences for qRT-PCR analysis.
Western blot assay
Proteins extracted from miR-515-5p mimics treated MGC-803 cells and miR-515-5p inhibitor-treated SGC-7901 cells were prepared by RIPA buffer followed by centrifugation under the conditions of 12,000 rpm/mins, 15 min. After determining the concentration of proteins with BCA kit, 40 μg of proteins were loaded and isolated by 8% SDS-PAGE. Subsequently, the targeted proteins were transferred onto nitrocellulose membranes (Millipore, Billerica, MA, USA). Low fat dried milk (5%) was used to incubate with membranes to block non-specific binding sites for 2 h. Then the membranes were incubated with primary antibodies that against XIAP (1:500, ab21278, Abcam) and GAPDH (1:10000, sc420485, Satna Cruz) overnight at 4 °C. Membranes were washed twice with PBS and then incubated with horseradish peroxidase-conjugated secondary antibodies for 2 h. Finally, the signals were examined by enhanced chemiluminescent reagents.
MTT assay
Cell viability of treated GC cells was examined by MTT dye reduction method. Treated GC cells were plated into 96-well plates and maintained at 37 °C for 24 h. Then, 20 μl dye solution was added into each well and incubated at 37 °C for more 4 h. Stop solution (100 μl) was then added into each well to stop reaction, and absorbance was determined at 570 nm.
Cell proliferation analysis
Effects of circ_0067997 and miR-515-5p on cell proliferation were evaluated by colony formation assay. In brief, treated MGC-803 or SGC-7901 cells were plated into 6-well plates at a density of 1000 cells/well and maintained at 37 °C for two weeks. Colonies were then fixed in methanol and stained with Giemsa, the number of colonies was calculated under a microscope.
Invasion analysis
Effects of circ_0067997 and miR-515-5p on cell invasion were assessed via trans-well assay using Matrigel matrix (BD Biosciences, USA) coated chambers with 8 µm pores (Corning Incorporated, Corning, USA). Briefly, treated GC cells were collected and re-suspended in 0.2 ml of culture medium, making the final concentration of 1 × 105/ml, and then added into the upper chamber. The lower chamber was filled with 0.5 ml of culture medium containing 10% FBS. After incubation for 24 h, the invaded GC cells were stained by 0.5% crystal violet (Beyotime Institute of Biotechnology, China), and counted with a microscope.
In vivo tumour formation assay
Six-week-old male BALB/c mice were utilized in the present study for in vivo tumour formation assay. Animal protocols in this study were approved by the Institutional Animal Care and Use Committee of the People’s Hospital of Anyang city. MGC-803 cells stably transfected with si-circ_0067997 or si-NC were collected and re-suspended in medium, making the final concentration of 1 × 106 cells/ml. Subsequently, 0.2 ml of treated MGC-803 cell suspension was injected into the right flank of nude mice. Tumour growth was detected every 7 days until 28 days after injection, and the volume of tumours was examined as the length × width2×0.5.
Plasmid construction and dual luciferase activity assay
In the recombinant plasmid construction, the fragments of circ_0067997 and XIAP 3′-UTR containing the complementary sequences of miR-515-5p, as well as their corresponding mutant sequences were amplified, respectively. The target fragments were then sub-cloned into the luciferase vector psi-CHECK (Promega, Madison, USA) to form circ_0067997 and XIAP 3′-UTR wild-type or mutant recombinant plasmids (circ_0067997-WT/Mut and XIAP 3′-UTR WT/Mut).
For dual luciferase reporter assay between circ_0067997 and miR-515-5p, MGC803 and SGC-7901 cells were plated into 24-well plates and maintained at 37 °C for 24 h, followed by co-transfecting circ_0067997-WT/Mut or XIAP 3′-UTR WT/Mut with miR-515-5p mimics or miR-515-5p inhibitor using Lipofectamine 2000 (Invitrogen). Dual-Luciferase Reporter Assay System (Promega) was utilized to detect the luciferase activity, and renilla luciferase activity was normalized to Firefly luciferase activity.
circRNAs immunoprecipitation (circRIP)
The interaction between circ_0067997 and miR-515-5p was validated by circRIP using the biotin-labelled circ_0067997 probe, which was designed and produced by Sangon Biotech, China. In brief, after fixing with 4% formaldehyde for 20 min, MGC-803 cells were lysed with RIPA buffer, followed by centrifugation under 12,000 g/min for 5 min. subsequently, 100 μl of the supernatants were retained as input and the remaining supernatants were incubated with the mixture of biotin-labelled circ_0067997 probe and streptavidin dynabeads (Invitrogen) overnight. The mixture was incubated with lysis buffer containing proteinase K, and then subjected to the RNA extraction and qRT-PCR detection.
Immunohistochemical analysis
For immunohistochemical of XIAP, tumour samples collected from xenografts formed by si-NC or si-circ_0067997 treated MGC-803 in BALA/c mice were fixed by 4% formalin and then cut into 4 μm sections. After dewaxing and rehydration, all sections were incubated for 5 min with 10 mM citrate buffer at 100 °C for antigen retrieval. After blocked non-specific binding sites, sections were incubated at 4 °C with rabbit polyclonal antibodies against XIAP (1:500, ab21278, Abcam) overnight. Subsequently, sections were subjected to the incubation with anti-rabbit secondary antibody at room temperature for 2 h, then the signals were detected by streptavidin-horseradish-peroxidase.
Statistical analysis
Data in the present study were expressed as mean ± SEM, GraphPad (Ver. Prism 7, GraphPad Prism Software, La Jolla, CA, USA) was utilized to perform statistical analysis using one-way analysis of variance or student t-test. Differences between means were considered significant if p values less than .05.
Results
Circ_0067997 was identified to be upregulated in GC
To screen circRNAs that involved in the tumorigenesis of GC, we first searched the GEO database and four GSE datasets (GSE78092, GSE83521, GSE89143 and GSE93541) were chosen to analyze the differently expressed circRNAs by GEO2R online tool. The differentially expressed cirRNAs were identified in the four GSE datasets by GEO2R online tool (). In order to get the more precise circRNAs differently expression profile, we modified the thresholds into p < .01 and FC >1.5, just one circRNA was identified in three GSE datasets, named as circ_0067997 (). Circ_0067997 arose from the FNDC3B gene and produced by the head-to-tail splicing of exon 8–11 (). We then detect the expression of circ_0067997 in GC cell lines and GES-1 cell line via qRT-PCR, results showed that circ_0067997 expression was significantly increased in four GC cell lines (SGC-7901, MGC-803, BGC-823, and MKN28) compared to that in GES-1 cell line (*p < .05, **p < .01, ). Expression of circ_0067997 in GC and matched non-cancer tissue samples from 48 patients were evaluated to further confirm its upregulation. As results indicated that circ_0067997 expression was remarkably upregulated in GC tissues compared to non-cancer tissues (***p < .001, ), and its expression was positively correlated with the TNM stages of GC (**p < .01, ). Moreover, GC patients with high circ_0067997 expression showed a worse overall survival rate than those with low circ_0067997 expression (*p < .05, ). In addition, we examined its parent gene FNDC3B expression in The Cancer Genome Atlas (TCGA) dataset by GEPIA online tool. The results revealed that the FNDC3B expression was also increased in GC tissues compared to normal tissues (*p < .05, ), and its expression also shown a tendency positively correlated with the TNM stages of GC (no statistically significant, ). In our cohort, we found that there was a positive correlation between circ_0067997 and FNDC3B expression in GC (*p < .05, ). Furthermore, the 48 GC patients were divided into high circ_0067997 expression group (n = 21) and low circ_0067997 expression group (n = 27), and statistical results showed that circ_0067997 expression was closely associated with GC TNM stages (p = .005) and differentiation grade (p = .016), but not age (p = .529), and gender (p = .399, ). Their data imply circ_0067997 may serve as a candidate for GC diagnose and treatment.
Figure 1. Circ_0067997 was identified to be upregulated in GC. (a) Differentially expressed circRNAs of GSE78092, GSE83521, GSE89143 and GSE93541 datasets were screened by online GEO2R tool based on different thresholds of P and FC. (b) The schematic diagram of genomic location of circ_0067997. (c–e) Relative expression level of circ_0067997 were assessed by qRT-PCR assay in GC cell lines (SGC-7901, MGC-803, BGC-823, MKN28) and GES-1 cell line, as well as in GC tissues and corresponding normal tissues. (f) The overall survival rate of GC patients with high or low expression of circ_0067997 was analyzed by Kaplan–Meier survival plots. (g and h) Relative mRNA expression of FNDC3B was examined in GC tissues from TCGA dataset qRT-PCR assay. (i) The correlation between circ_0067997 and FNDC3B expression in GC tissues. *p < .05, **p < .01, ***p < .001.
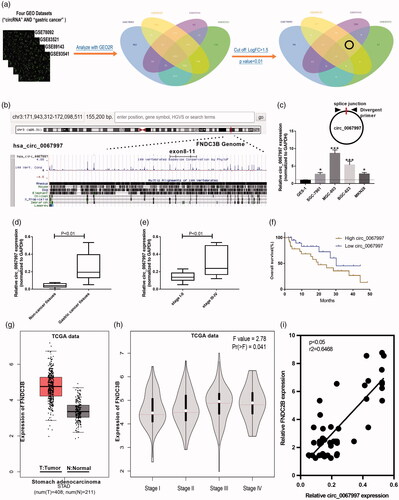
Table 2. Correlations between circ_0067997 expression and clinicopathologic characteristics in GC.
Circ_0067997 promoted GC progression in vitro and in vivo
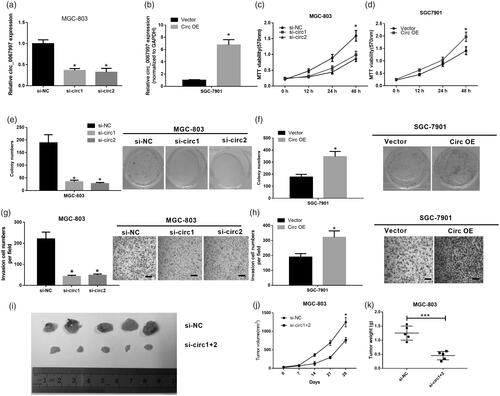
We then blocked the circ_0067997 expression in MGC-803 cells (higher expression) by its siRNAs and overexpressed circ_0067997 expression in SGC7901 cells(lower expression) by treating cells with circ_0067997, to explore the roles of circ_0067997 in GC progression. The efficiency of circ_0067997 knockdown and overexpression were evaluated by qRT-PCR, MGC-803 cells transfected with circ_0067997 siRNAs showed a significant downregulation of circ_0067997 than those cells transfected with si-NC (*p < .05, ); SGC-7901 cells transfected with circ_0067997 showed a remarkable upregulation of circ_0067997 compared to those cells treated with empty vector (*p < .05, ). Subsequently, the effects of circ_0067997 on cell viability, proliferation and invasion were assessed by MTT, colony formation, and trans-well assays, respectively. MTT results indicated that knockdown of circ_0067997 in MGC-803 cells significantly reduced cell viability (**p < .01, ), whereas overexpression of circ_0067997 in SGC-7901 cells remarkably increased cell viability (**p < .01, ). Decreased circ_0067997 expression in MGC-803 cells suppressed the colony formation ability (**p < .01, ), whereas increased circ_0067997 expression in SGC-7901 cells promoted the colony formation ability (**p < .01, ). Similarly, circ_0067997 silenced MGC-803 cells showed a markedly attenuated invasive ability (***p < .001, ), whereas circ_0067997 overexpressed SGC-7901 cells exhibited a remarkably enhanced invasive ability (***p < .001, ). In addition, in vivo tumour growth assay was performed to further investigate the roles of circ_0067997. MGC-803 cells stably transfected with circ_0067997 siRNAs or si-NC were subcutaneously injected into the left flank of BALA/c mice and allowed to grow for 28 days. Results indicated that volume and weight of tumours from circ_0067997 siRNAs treated MGC-803 cells were significantly reduced compared with those tumours from si-NC treated cells (*p < .05, ).
Figure 2. Circ_0067997 promoted GC progression in vitro and in vivo. (a) Relative expression level of circ_0067997 of MGC-803 cells transfected with circ_0067997 siRNAs and its negative control (si-NC). (b) Relative expression of circ_0067997 of SGC-7901 cells transfected with empty vector and circ_0067997. (c and d) MTT assay was utilized to examine cell viability of circ_0067997 blocked MC-803 cells and circ_0067997 overexpressed SGC-7901 cells. (e and f) Colony formation was carried out to detect cell proliferation of circ_0067997 blocked MGC-803 cells and circ_0067997 overexpressed SGC-7901 cells. (g and h) Trans-well assay was performed to assess cell invasion of circ_0067997 blocked MC-803 cells and circ_0067997 overexpressed SGC-7901 cells. (i-k) Hypodermic injection of circ_0067997 siRNAs and si-NC treated MGC-803 cells into BALB/c nude mice to construct xenograft model, tumour volume was measured every 7 days until 28 days, tumour weight was assessed at 28 days after transfection. *p < .05, **p < .01, ***p < .001.
Circ_0067997 served as a sponge for multiple miRNAs
Since circRNAs were demonstrated to act as sponge for miRNAs in previous studies, we next explored the potential miRNAs associated with circ_0067997 by circularRNA interactome prediction. Results showed that circ_0067997 might be the potential sponges for 17 miRNAs (). We then investigated whether these miRNAs could be regulated by circ_0067997. qRT-PCR was used to detect miRNAs that in circ_0067997 silenced MGC-803 and circ_0067997 overexpressed SGC-7901 cells. The results showed five miRNAs (miR-127–5, miR-451, miR-515-5p, miR-615-5p, and miR-625) were significantly changed (*p < .05, **p < .01, ***p < .001, ). Subsequently, the targeted genes of these five miRNAs were screened by Targetscan7.2 and then analyzed via KEGG. The most significant pathway involved in miR-127–5, miR-451, miR-515-5p, miR-615-5p and miR-625 expression were shown, among which we found several GC related pathways, such as hsa04310: Wnt signalling pathway, hsa04115: p53 signalling pathway, hsa04210: Apoptosis, hsa04150: mTOR signalling pathway, hsa04310: Wnt signalling pathway, and hsa03320: PPAR signalling pathway (). Supposing that circ_0067997 might play a role in GC through ceRNA mechanism. Then circRNA-miRNA-target genes network of circ_0067997 was established within its five targeted miRNAs ().
Figure 3. Circ_0067997 served as a sponge for multiple miRNAs. (a and b) Relative expression levels of 17 miRNAs sponged by circ_0067997 were evaluated in circ_0067997 blocked MGC-803 and circ_0067997 overexpressed SGC-7901 cells. (c) The targeted genes of miR-127–5, miR-451, miR-515-5p, miR-615-5p, and miR-625 were predicted by Targetscan7.2 and then analyzed via KEGG, respectively. (d) The circRNA-miRNA-mRNA regulatory network of circ_0067997 was established within its five targeted miRNAs. *p < .05, **p < .01, ***p < .001.
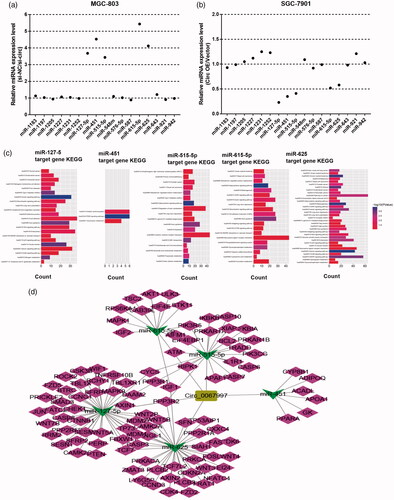
Table 3. Circular RNA interactome prediction of miRNAs binding sites with circ_0067997.
Circ_0067997 negatively regulates miR-515-5p by directly binding to it
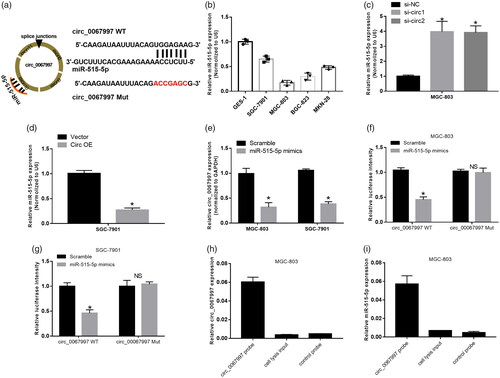
According to the prediction of circular RNA interactome, circ_0067997 possessed complementary sequence to miR-515-5p seed region (). Results from qRT-PCR showed that miR-515-5p expression was significantly decreased in GC cell lines compare to GES-1 cell line (*p < .05, ). Knockdown of circ_0067997 in MGC-803 cells remarkably increased miR-515-5p expression (*p < .05, ), whereas overexpression of circ_0067997 in SGC-7901 cells significantly decreased miR-515-5p expression (*p < .05, ). The expression of circ_0067997 of MGC-803 and SGC-7901 cells treated with miR-515-5p mimics were markedly lower than those cells treated with scramble control (*p < .05, ). Luciferase reporter assay was carried out to validate the direct interaction between circ_0067997 and miR-515-5p in MGC-803 and SGC-7901 cells, miR-515-5p mimics significantly attenuated the luciferase activity driven by circ_0068997 WT, but not the luciferase activity driven by circ_0068997 Mut (*p < .05, ). In addition, RNA precipitation (RIP) was used to further confirm the interaction between circ_0067997 and miR-515-5p in MGC-803 and SGC-7901 cells using circ_0068997 specific probe. Associated RNAs pulled down by circ_0068997 specific probe were purified and detected using qRT-PCR. As excepted, we revealed a specific enrichment of circ_0068997 and miR-515-5p compare to control probe (***p < .001, ).
Figure 4. Circ_0067997 directly bonded to miR-515-5p and negatively regulated miR-515-5p. (a) The putative wild-type and mutant type binding site sequence of miR-515-5p in circ_0067997. (b) Relative expression of miR-515-5p was assessed in GC cell lines and GSE-1 cell line by qRT-PCR. (c and d) Expression level of miR-515-5p in circ_0067997 siRNAs treated MGC-803 and circ_0067997 recombinant plasmid treated SGC-7901 cells were examined by qRT-PCR. (e) Relative expression of circ_0067997 of miR-515-5p treated MGC-803 and SGC-7901 cells were detected by qRT-PCR. (f and g) Interaction between circ_0067997 and miR-515-5p was validated by luciferase reporter assay using recombinant luciferase plasmid driven by circ_0067997 WT or circ_0067997 Mut in MGC-803 and SGC-7901 cells. (h and i) Circ_0067997 in MGC-803 and SGC-7901 cell lysis were pulled down by specific probe and detected by qRT-PCR. *p < .05, **p < .01, ***p < .001.
miR-515-5p targets and negatively regulates XIAP and suppressed GC cell proliferation
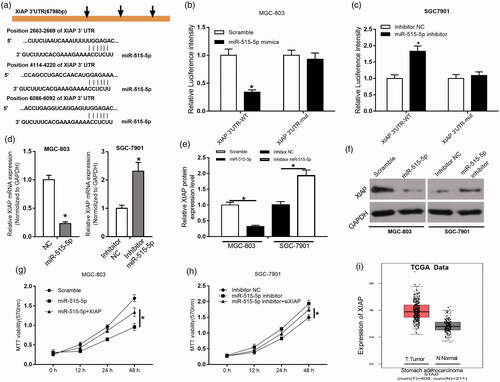
To further elucidate the biological mechanisms underlying the roles of miR-515-5p in GC, we investigated the potential target genes of miR-515-5p. According to the prediction results, miR-515-5p could target XIAP 3′-UTR with a high score, and XIAP possessed three complementary sequences to miR-515-5p (). In the luciferase reporter assay, we found that miR-515-5p could suppress the luciferase activity driven by XIAP 3′-UTR-WT, but boot XIAP 3 ′ -UTR-Mut in MGC-803 and SGC-7901 cells (*p < .05, ). Relative mRNA expression of XIAP was significantly reduced in miR-515-5p mimics treated MGC-803 cells compared to those cells treated with NC, whereas it was remarkably increased in miR-515-5p inhibitor-treated SGC-7901 cells compared with those cells treated with inhibitor-NC (*p < .05, ). Western blot assay showed a similar trend of XIAP protein expression with its mRNA expression in miR-515-5p mimics treated MGC-803 cells and miR-515-5p inhibitor-treated SGC-7901 cells (*p < .05, ). In the MTT assay, we found that miR-515-5p treatment could reduce MGC-803 and SGC-7901 cell viability, however, these suppressive effects of miR-515-5p on cell viability are abolished by the application of XIAP (*p < .05, ). In addition, we examined XIAP expression profiling dataset from TCGA by GEPIA online tool, results indicated that XIAP expression was remarkably upregulated in GC tissues compared to normal tissues (*p < .05, ).
Figure 5. miR-515-5p targeted and negatively regulated XIAP and suppressed GC cell proliferation. (a) The putative sequences of miR-515-5p and XIAP 3′-UTR with three binding sites. (b and c) Interaction between miR-515-5p and XIAP was validated by luciferase reporter assay in MGC-803 and SGC-7901 cells. (d–f) Relative mRNA and protein expression of XIAP in miR-515-5p mimics treated MGC-803 and miR-515-5p inhibitor-treated SGC-7901 cells were examined by qRT-PCR and western blot, respectively. (g and h) Cell proliferation of miR-515-5p mimics treated MGC-803 and miR-515-5p inhibitor-treated SGC-7901 cells were determined by MTT assay with the presence of XIAP or not. (i) Expression of XIAP was analyzed in GC and normal tissues from TCGA dataset. *p < .05, **p < .01, ***p < .001.
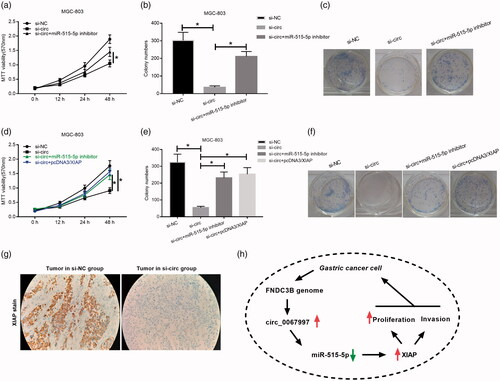
XIAP and miR-515-5p inhibitor could reverse the effects of circ_0067997 knockdown on MGC-803 cells
After validating that XIAP was a direct target gene of miR-515-5p, we sought to determine whether the effects of circ_0067997 on GC cells were miR-515-5p/XIAP axis dependent. In functional assays, we found that miR-515-5p inhibitor could reverse the suppressive effects of circ_0067997 knockdown on cell viability and proliferation (*p < .05, ). Similarly, we found that pcDNA/XIAP could also abolish the suppressive effects of circ_0067997 knockdown on cell viability, and proliferation (*p < .05, ). In addition, we analyzed the XIAP expression in tumour sections collected from in vivo xenograft by immunohistochemical staining, results showed that XIAP expression was significantly reduced in si-circ_0067997 treated tumour group compared to si-NC treated group (). Taken together, increased circ_0067997, formed by FNDC3B, could upregulate the expression of XIAP by inhibiting miR-515-5p and then promote GC cell proliferation and invasion ().
Figure 6. XIAP and miR-515-5p inhibitor could reverse the effects of circ_0067997 knockdown on MGC-803 cells. (a) Effects of miR-515-5p inhibitor treatment on cell viability of circ_0067997 knockdown MGC-803 cells were evaluated by MTT assay. (b and c) Effects of miR-515-5p inhibitor treatment on cell proliferation of circ_0067997 blocked MGC-803 cells were examined by colony formation assay. (d) Effects of XIAP treatment on cell viability of circ_0067997 knockdown MGC-803 cells were evaluated by MTT assay. (e and f) Effects of XIAP application on cell proliferation of circ_0067997 blocked MGC-803 cells were examined by colony formation assay. (g) Immunohistochemical staining in tumour slice from in vivo xenograft was performed to determine the expression of XIAP. (h) The molecular mechanisms of circ_0067997 in the tumorigenesis of GC.
Discussion
GC is now become a serious public health issue, affecting more than millions of people in the world [Citation16]. Despite GC usually develops in stages over several years, it still progresses in the majority of GC patients to advanced stages, which characterized by distant metastasis and poor prognosis [Citation17]. In most cases, advanced GC is inoperable because of its wide metastasis, and the current therapy for GC patients is mainly dependent on systemic chemotherapy [Citation18]. However, chemo-resistance of GC significantly limits the effects of chemotherapy, reducing the survival rates of patients [Citation19]. Although increasing studies devote to understand the tumorigenesis of GC, the associated mechanisms remain largely unclear. Our present study indicated a novel molecule that could act as an oncogene in GC progression, and we revealed a new pathway, which may serve as a diagnose or treatment target for GC.
Abnormal circRNAs expression profile was revealed closely linked to the initiation and progression of multiple human cancers, including colon cancer, breast cancer and lung cancer [Citation20–22]. Various circRNAs have been applied as biomarkers to evaluate the viability, metastasis, drug-resistance, invasion and migration of different cancer cells [Citation23]. For example, circ_0072995 was identified to be upregulated in breast cancer cells, and its expression showed a positive correlation with the cell invasion and migration [Citation24]. Xing J et al. demonstrated that circ_CEP128 expression was significantly increased in bladder cancer samples compared to normal samples, and blocked its expression, would induce the suppression of cell proliferation and the promotion of cell apoptosis [Citation25]. Although the exact molecular mechanisms of GC tumorigenesis remain largely undetermined, emerging evidences indicated that circRNAs expression profiles of GC exhibited dysregulation [Citation26]. Moreover, multiple circRNAs were revealed to play critical roles in GC cell proliferation, apoptosis and invasion [Citation27]. For instance, circ_PVRL3 expression was found to be downregulated in GC samples when compared with adjacent normal samples, and decreased circ_PVRL3 expression was contributed to the proliferation and invasion of GC cells [Citation28]. These findings indicated that circRNAs involved in the progression of GC, suggesting that circRNAs may be a potential therapeutic target for GC. In our study, circ_0067997 was identified to be upregulated in four GSE datasets using circRNAs microarray, which was validated by qRT-PCR in GC tissues and cell lines. GC patients with high circ_0067997 expression showed a worse overall survival rate than those with low circ_0067997 expression. Moreover, in vitro and in vivo functional assays indicated that circ_0067997 exhibited promotive effects on GC cell viability, proliferation and invasion. These findings suggested that circ_0067997 involved in the progression of GC, and it might be a new effective therapeutic target of GC.
It has been well documented that circRNAs participated in the regulation of multiple physiological processes by acting as sponge of miRNAs [Citation29]. Moreover, the miRNA-mRNA axis was frequently revealed to be activated or suppressed in multiple tumour cells by circRNAs, exhibiting positive or negative effects on cancer progression [Citation29]. For instance, Zhang L et al. reported that circRNA_ACAP2 could promote cell proliferation, invasion and migration of colon cancer cells through miR-21-5p/Tiam1 regulatory circuit [Citation30]. In the present study, circular RNA interactome was used to predict the target miRNAs of circ_0067997, and qRT-PCR was utilized to screen the miRNAs among them that could be regulated by circ_0067997. We validated that circ_0067997 serve as a sponge of miR-515- 5p in GC cells.
XIAP, involved in various biological processes, is an apoptotic regulator that was identified by Liston [Citation31]. XIAP can block many apoptosis-related pathways and, therefore, it was reported to be an attractive therapeutic target of multiple human tumors [Citation32]. In addition, the effects of miRNAs on tumour progression were reported to be mediated through regulation of XIAP expression by directly binding to XIAP [Citation33,Citation34]. In the present study, XIAP was identified as a target gene of miR-515-5p, and could be regulated by circ_0067997. In the functional assays, miR-515-5p inhibitor and pcDNA3/XIAP abolished the effects of circ_0067997 knockdown on GC cells. These results revealed a new pathway in GC progress.
In conclusion, we have these major findings in the present study: (1) We reported circular RNA, hsa_circ_0067997 in gastric cancer. (2) We studied miR-515-5p in gastric cancer progress. (3) We reported circ_0067997 sponges miR-515-5p in gastric cancer cells. (4) MiR-515-5p directly targets XIAP in gastric cancer cells. (5) Circ_0067997/miR-515-5p/XIAP axis plays critical role in gastric cancer pathogenesis.
Disclosure statement
The authors have no commercial or other associations that might pose a conflict of interest.
Additional information
Funding
References
- Hamashima C. Current issues and future perspectives of gastric cancer screening. Wjg. 2014;20:13767–13774.
- Ferlay J, Colombet M, Soerjomataram I, et al. Cancer incidence and mortality patterns in Europe: estimates for 40 countries and 25 major cancers in 2018. Eur J Cancer. 2018.Available from: https://doi.org/10.1016/j.ejca.2018.07.005
- Herrero R, Park JY, Forman D. The fight against gastric cancer - the IARC Working Group report. Best Pract Res Clin Gastroenterol. 2014;28:1107–1114.
- Kimura T, Egawa S. Epidemiology of prostate cancer in Asian countries. Int J Urol. 2018;25:524–531.
- Coccolini F, Montori G, Ceresoli M, et al. Advanced gastric cancer: what we know and what we still have to learn. Wjg. 2016;22:1139–1159.
- Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12:381–388.
- Wang PL, Bao Y, Yee MC, et al. Circular RNA is expressed across the eukaryotic tree of life. PLoS One. 2014;9:e90859.
- Shin VY, Chu KM. MiRNA as potential biomarkers and therapeutic targets for gastric cancer. Wjg. 2014;20:10432–10439.
- Song H, Xu Y, Shi L, et al. LncRNA THOR increases the stemness of gastric cancer cells via enhancing SOX9 mRNA stability. Biomed Pharmacother. 2018;108:338–346.
- Qu D, Yan B, Xin R, et al. A novel circular RNA hsa_circ_0020123 exerts oncogenic properties through suppression of miR-144 in non-small cell lung cancer. Am J Cancer Res. 2018;8:1387–1402.
- Chen F, Feng Z, Zhu J, et al. Emerging roles of circRNA_NEK6 targeting miR-370-3p in the proliferation and invasion of thyroid cancer via Wnt signaling pathway. Cancer Biol Ther. 2018; 1–14.
- Xiao B, Wen J, Zhao C, et al. Differences in expression profiles of circular RNA between luminal breast cancer cells and normal breast cells. Nan Fang Yi Ke Da Xue Xue Bao. 2018;38:1014–1019.
- Dang Y, Ouyang X, Zhang F, et al. Circular RNAs expression profiles in human gastric cancer. Sci Rep. 2017;7:9060.
- Shen Y, Zhang J, Fu Z, et al. Gene microarray analysis of the circular RNAs expression profile in human gastric cancer. Oncol Lett. 2018;15:9965–9972.
- Huang YS, Jie N, Zou KJ, et al. Expression profile of circular RNAs in human gastric cancer tissues. Mol Med Rep. 2017;16:2469–2476.
- Lin MT, Song HJ, Ding XY. Long non-coding RNAs involved in metastasis of gastric cancer. World J Gastroenterol. 2018;24:3724–3737.
- Harada K, Lopez A, Shanbhag N, et al. Recent advances in the management of gastric adenocarcinoma patients. F1000Res. 2018;7:F1000 Faculty Rev-1365.
- Magalhaes H, Fontes-Sousa M, Machado M. Immunotherapy in advanced gastric cancer: an overview of the emerging strategies. Can J Gastroenterol Hepatol. 2018;2732408. Available from: https://doi.org/10.1155/2018/2732408
- Bolm L, Kasmann L, Paysen A, et al. Multimodal anti-tumor approaches combined with immunotherapy to overcome tumor resistance in esophageal and gastric cancer. Anticancer Res. 2018;38:3231–3242.
- Yuan G, Chen T, Zhang H, et al. Comprehensive analysis of differential circular RNA expression in a mouse model of colitis-induced colon carcinoma. Mol Carcinog. 2018;57(12):1825-1834.
- Shi P, Sun J, He B, et al. Profiles of differentially expressed circRNAs in esophageal and breast cancer. Cancer Manag Res. 2018;10:2207–2221.
- Hang D, Zhou J, Qin N, et al. A novel plasma circular RNA circFARSA is a potential biomarker for non-small cell lung cancer. Cancer Med. 2018;7:2783–2791.
- Zhang HD, Jiang LH, Sun DW, et al. CircRNA: a novel type of biomarker for cancer. Breast Cancer. 2018;25:1–7.
- Zhang HD, Jiang LH, Hou JC, et al. Circular RNA hsa_circ_0072995 promotes breast cancer cell migration and invasion through sponge for miR-30c-2-3p. Epigenomics. 2018;10(9):1229-1242.
- Wu Z, Huang W, Wang X, et al. Circular RNA CEP128 acts as a sponge of miR-145-5p in promoting the bladder cancer progression via regulating SOX11. Mol Med. 2018;24:40.
- Shao Y, Li J, Lu R, et al. Global circular RNA expression profile of human gastric cancer and its clinical significance. Cancer Med. 2017;6:1173–1180.
- Li J, Yang J, Zhou P, et al. Circular RNAs in cancer: novel insights into origins, properties, functions and implications. Am J Cancer Res. 2015;5:472–480.
- Sun HD, Xu ZP, Sun ZQ, et al. Down-regulation of circPVRL3 promotes the proliferation and migration of gastric cancer cells. Sci Rep. 2018;8:10111.
- Chan JJ, Tay Y. Noncoding RNA: RNA regulatory networks in cancer. Int J Mol Sci. 2018;19:1310-1336.
- He JH, Li YG, Han ZP, et al. The CircRNA-ACAP2/Hsa-miR-21-5p/Tiam1 regulatory feedback circuit affects the proliferation, migration, and invasion of colon cancer SW480 cells. Cell Physiol Biochem. 2018;49:1539–1550.
- Lencz T, Guha S, Liu C, et al. Genome-wide association study implicates NDST3 in schizophrenia and bipolar disorder. Nat Commun. 2013;4:2739.
- Sun J, Li J, Zhang W, et al. MicroRNA-509-3p inhibits cancer cell proliferation and migration via upregulation of XIAP in gastric cancer cells. Oncol Res. 2017;25:455–461.
- Liu S, Zhang P, Chen Z, et al. MicroRNA-7 downregulates XIAP expression to suppress cell growth and promote apoptosis in cervical cancer cells. FEBS Lett. 2013;587:2247–2253.
- Gu C, Wang Z, Jin Z, et al. MicroRNA-212 inhibits the proliferation, migration and invasion of renal cell carcinoma by targeting X-linked inhibitor of apoptosis protein (XIAP). Oncotarget. 2017;8:92119–92133.
