Abstract
Presently, clopidogrel is the standard therapeutic drugs for antiplatelet therapy. Variants in the CYP2C19 gene influence the clinical response of clopidogrel. Thus, the US Food and Drug Administration suggested CYP2C19 genotyping needs to identify before taking medicine. Due to high cost, time consuming, and sophisticated instruments, current single nucleotide polymorphism detection methods are limited in clinical application. In the present study, we established a genotyping method for CYP2C19, which combines amplification refractory mutation system (ARMS)-PCR with a lateral flow assay used gold magnetic nanoparticles (GMNPs) named as PCR-gold magnetic lateral flow assay system (PCR-GoldMag LFA). The PCR products with specific genotype can be explained within 5 minutes, either through visual or by a magnetic reader automatically according to the captured GMNPs probes on the test lines of the LFA strips. The limit of detection of this method is 5 ng of genomic DNA. The PCR-GoldMag LFA system was applied in a clinical trial with 1356 samples of Han Chinese. The concordance rate between the LFA system and sequencing is 99.93%. The allele frequency of CYP2C19*2 and CYP2C19*3 are 30.38 and 7.08% in Han Chinese, respectively. This method provides a new way in the clinical application of CYP2C19 genotyping to guide the clopidogrel medication.
Introduction
Presently, clopidogrel (alone or in association with aspirin) is the standard therapeutic drugs for antiplatelet therapy after acute coronary syndromes and percutaneous coronary intervention. Unfortunately, after about 25% of the patients treated with the standard clopidogrel dosage, a poor clinical effect is obtained sometimes leading to serious adverse reactions Including myocardial infarct, stroke, or death have been described [Citation1,Citation2]. Cytochrome P450 (CYP) 2C19 is an enzyme involved in the bioactivation of various drugs. Variants in the CYP2C19 gene influence the pharmacokinetics and clinical response to antiplatelet drugs such as clopidogrel [Citation3]. The CYP2C19*2 (c.681G>A rs4244285) and the CYP2C19*3 (c.636G>A rs4986893) alleles are the most common and investigated polymorphism. Carriers of at least one loss-of-function allele have a reduced response to clopidogrel and thus a higher risk for platelet aggregation when compared with noncarriers after taking the drug [Citation4]. Therefore, the US Food and Drug Administration (FDA) suggested that it is necessary to identify the genotype status prior to prescribing clopidogrel.
Among the masses of current single nucleotide polymorphism (SNP) detection methods, direct sequencing is regarded as the golden standard for the detection of SNP mutations. However, according to the methodology theory and the subsequent massive data analysis, sequencing is more suitable for the discovery of novel SNPs [Citation5–7] than direct detection of specifically known SNPs. Other technologies, such as pyrosequencing [Citation8], denatured high-performance liquid chromatography (dHPLC) [Citation9], and Mass ARRAY [Citation10] have been used to detect various types of mutations. More recently, several new techniques, such as RT-PCR [Citation11], PCR-RFLP [Citation12], and high-resolution melting (HRM) technique [Citation7] also offered sensitive methods to detect SNPs. However, the application of these techniques for clinical is still limited because they are relatively expensive, time consuming, or invariably require favorable experimental conditions and sophisticated instruments [Citation13,Citation14]. The tremendous demands of CYP2C19 genotyping in clinical practice had to better meet the requirements of point-of-care testing (POCT) strategies [Citation15].
Gold magnetic nanoparticles are Fe3O4/Au/Fe3O4 composite particles with significantly elevated stableness and compatibility for biological interactions [Citation16,Citation17]. It shows great potential for serving as diagnostic or therapeutic agents in a variety of medical fields [Citation18–20]. In a previous study, our laboratory established a novel lateral flow assay (LFA) system assembled with GMNPs for the visual detection of MTHFR C677T polymorphism which relies on the hybridization reaction on test line [Citation21]. This method is simple, accurate, affordable, and user-friendly for SNP detection. Due to these advantages, we hope the method can be applied to CYP2C19 genotyping.
A rapid and accurate method for CYP2C19 genotyping was proposed. The result can be explained by a magnetic reader or visual check. This method simplifies the procedure of genotyping and avoids sophisticated instruments to satisfy the application in clinical practice [Citation22]. The accuracy of this system was confirmed with DNA sequencing by 1356 clinical samples of Han Chinese. In addition, this clinical trial also provided information about the distribution of CYP2C19 gene polymorphisms in a large Chinese Han population.
Materials and methods
Reagent
HotMaster Taq DNA polymerase was obtained from TIANGEN Biotech Co., Ltd. (Beijing, China). dNTP and uracil-DNA glycosylase (UDG) polymerase were purchased from ShineGene Molecular Biotechnology Co., Ltd. (Shanghai, China). Anti-digoxin antibody was from Meridian Life Science, Inc. (Saco, ME, USA) and Streptavidin from Promega Biotech Inc. (Madison, WI, USA). Goat anti-mouse IgG was purchased from Joey Bioscience Inc. (Shanghai, China). GMNPs and lateral flow strips were from Xi’an GoldMag Nanobiotech Co., Ltd (Xi’an, China). All primers were synthesized by Invitrogen Biotechnology Ltd. (Shanghai, China).
Plasmid construction and identification
The genomic DNA of CYP2C19*2 hybrid type and CYP2C19*3 wild type which were sequenced by Sangon Biotech Co., Ltd. (Shanghai, China) as a template, using the primer pairs *2-seq-F/*2-seq-R and *3-seq-F/*3-seq-R () for amplification and then obtained the amplification products of target fragment. CYP2C19*3 mutations were introduced by the site-directed mutagenesis method (primers in ). The plasmids were constructed using the pMD19-T Vector Cloning Kit (Takara Bio Inc. Dalian, China) with the amplification products of target fragment and were extracted from transformed JM 109 cells. The data indicated that four plasmids were successfully constructed and validated by sequencing (Figure S1).
Table 1. Primer sequences used for DNA sequencing, site-directed mutagenesis.
PCR amplification
According to the ARMS-PCR [Citation23,Citation24] for CYP2C19 genotyping, the forward primers were designed as the common one and the reverse primers are allele-specific primers because the nucleotide at the 3' terminal corresponds to the SNP site. To ensure the specificity of the primers, an additional mismatch at the penultimate or antepenultimate nucleotide of the 3' terminus of reverse primers (listed in ) was introduced [Citation25].
Table 2. Primer sequences used for ARMS-PCR.
After PCR amplification with these primers, the length of the CYP2C19*2 and *3 amplification products are 143 bp and 146 bp, respectively.
CYP2C19 alleles (*2 and *3) were detected separately by complementary reactions (M tube and WT tube). Each PCR reaction, whose volume was 50 μL in total, includes 10× PCR buffer (10 mM Tris HCl and 50 mM KCl), 0.2 mM dNTP mixture (dATP, dUTP, dCTP, and dGTP), 3 mM MgCl2, 0.5 U of Hotmaster Taq DNA polymerase, 0.5 U of UDG polymerase, 100 nM common forward primer and 3 μL genomic DNA. 100 nM of the reverse (M) primer in the M tube and the reverse (WT) primer in the WT tube were used separately. Applied Biosystems 2720 PCR Thermal Cycler (Applied Biosystems, Foster City, U.S.A.) was used to amplify the target fragment which was started as a UDG incubation step (50°C for 2 min) and a UDG inactivation step (95°C for 5 min), followed by 28 cycles of 94°C for 30 s, 60°C for 30 s, and 65°C for 1 min, and 65°C for 10 min as a final extension.
GoldMag LFA system
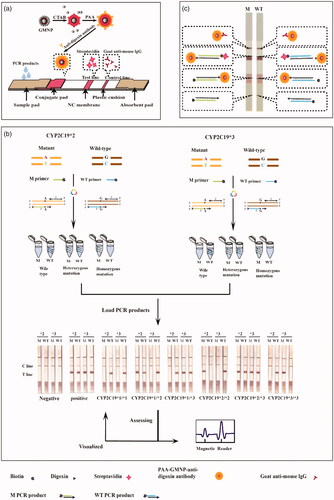
LFA strips contain five sections: support plate, sample pad, conjugate pad, nitrocellulose membrane, absorbent paper. Goat anti-mouse IgG and streptavidin were immobilized onto porous nitrocellulose (NC) membrane, respectively to form a control line (C-line) and a test line (T-line) by a BioJet (HM3010, BioDot, Inc., Irvine, CA, USA). GMNPs were synthesized [Citation17,Citation26] and modified according to the methods described previously. The GMNPs were firstly treated by cetyltrimethylammonium bromide (CTAB) surfactant and then modified by polyacrylic acid (PAA) [Citation16]. The GMNPs coupled with anti-digoxin antibody was fixed onto the conjugate pad of GoldMag LFA strips ().
Clinical applications
In this multi-center study, a total of 1356 samples of human blood were obtained from three hospitals including 414 specimen from Xiangya Hospital Central-South University (Changsha, China), 306 cases from Hunan Provincial People’s Hospital (Changsha, China), and 636 samples from Shaanxi Provincial People’s Hospital (Xi’an, China). These samples of Chinese Han populations were all from outpatient. All specimens collected approval of the hospital ethics committee. Genomic DNA was isolated from 100 μL EDTA blood using a whole blood genomic DNA isolation kit from Xi’an GoldMag Nanobiotech Co., Ltd (Xi’an, China) according to the manufacturer’s instructions. CYP2C19 genotyping was performed by the PCR-GoldMag LFA system. The DNA sequencing by Beijing Genomics Institute (BGI, Shenzhen, China) was used as a control method.
Statistical analysis
The Hardy–Weinberg equilibrium was assessed for each SNP to validate data reliability.
Categorical variables were represented as counts and percentages. Kappa test was used to analyze the consistency of the novel method with the gold standard. It is considered more consistent if the Kappa value was greater than 0.75. Chi-square test was used to test for differences between groups of categorical variables. Analyses were performed using SPSS version 19.0 statistical software (IBM Co., Armonk, NY, USA). A value of p < .05 was considered statistically significant.
Results and discussion
Principle of the PCR-GoldMag LFA system
The method for CYP2C19 genotyping was proposed which combines (ARMS)-PCR with a lateral flow assay. CYP2C19*2 and CYP2C19*3 were amplificated by ARMS-PCR simultaneously. Each SNP (CYP2C19*2 or *3) have two sets of primers, including common primer and allele-specific primers (corresponding M and WT), were respectively added into two tubes (M tube and WT tube) for PCR separately. After PCR amplification, the whole PCR products in four tubes (CYP2C19*2 and *3) were added onto the sample pad, respectively. The genotyping result can be read within 5 min through a magnetic reader automatically or be visually checked ().
When the target PCR products labelled both biotin and digoxin were added to the sample pads of GoldMag LFA strips, the digoxin labeled on the 5′-end affinity reacted with the anti-digoxin antibody-coated GMNPs on the conjugate pad and form a GoldMag-anti-digoxin antibody – digoxin labelled PCR product complex. The complex is driven by capillary force along the strip and then the biotin labeled on the 5′-end was fixed and enriched by immobilized streptavidin on the test line (T line), which make a red band at the T line due to the bathochromic effect of GMNPs. The goat anti-mouse IgG on the control line (C line) capture the rest of the complex, which confirms the efficacy of the lateral flow system (). For wild-type samples, the distinct red bands are only visible on the T-line of the WT channel, while the T-line of the M channel does not have color bands. On the contrary, for homozygous mutant samples, red bands appeared only on the M channel strips. However, when there are red bands with similar intensity on the T lines of the M and WT strips, it represents a heterozygous mutant sample.
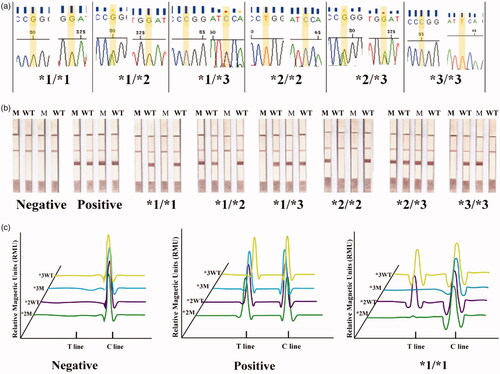
The accuracy of the LFA system was evaluated. Six CYP2C19 genotypes (*1/*1, *1/*2, *1/*3, *2/*2, *2/*3, and *3/*3), which have been validated by DNA sequencing () were evaluated by the PCR-GoldMag LFA system (). Based on the magnetic signals of GMNPs, the results were also detected by a magnetic reader (shown in and Figure S2). The PCR system without template was set as a negative control and the constructed plasmid (10 pg/μL) was used for the positive control. The result showed that the PCR-GoldMag LFA system can clearly distinguish the wild-type target DNA from the mutant target DNA by one base mutation, performing successfully the distinction between the CYP2C19 genotypes.
Figure 2. Accuracy of the PCR-GoldMag LFA system. (a) Six CYP2C19 genotypes were confirmed by DNA sequencing. (b) Six CYP2C19 genotypes were screened out by the PCR-GoldMag LFA system. (c) Based on the magnetic characteristics of GMNP, the results (negative, positive, and CYP2C19 *1/*1) were read by magnetic reader.
The whole process can be completed within 2 h. It needs 5 min to obtain the genotyping visually or magnetic reader for automatic inspection of colors on the T and C lines. Compared with the conventional PCR-based SNPs detection methods or DNA sequencing, the major advantage of this system is a rapid qualitative answer in “yes” or “no” terms. Based on the magnetic signal of GMNPs, these results can also be detected by a magnetic reader.
Optimization of the PCR-GoldMag LFA system for CYP2C19 genotyping
To ensure the specificity of the primers, three different pairs of mismatch were introduced and evaluated by genomic DNA of CYP2C19*2 hybrid type and CYP2C19*3 wild type. The results of primer screening were shown in Figure S3. It can be seen that the mismatch of medium strength at the penultimate nucleotide of the 3' terminus of allele-specific reverse primers is best.
We optimized a series of PCR systems including the concentration of primers, annealing temperature, and the PCR cycles. To determine the optimal concentration of the primers, different concentrations (150, 100, 50, 25 nM) of each primer were added to the PCR mixture. The results showed that 100 nM primers were optimal for CYP2C19 genotyping (Figure S4). We also tested the annealing temperatures (e.g. 56, 58, 60, 62 °C) and PCR cycles (e.g. for up to 26, 28, 30, 32 cycles). In order to increase high sensitivity without background, the best-optimized condition for PCR reaction is 58 °C of the annealing temperature (Figure S5) and 30 of the cycle number (Figure S6).
Performance of the PCR-GoldMag LFA system for CYP2C19 genotyping
The specificity was the most important factors for the PCR-GoldMag LFA System. In order to verify the specificity of this system for CYP2C19 genotyping, four plasmids were successively added in the *2 M PCR mixture in four tubs and they were added in the *2WT, *3M, *3WT PCR mixture the same as *2 M. The result showed that the PCR systems of *2 M, *2WT, *3 M, and *3WT can only amplify *2AA, *2GG, *3AA, and *3GG, respectively and cannot amplify other templates (). Therefore, PCR-GoldMag LFA system can clearly distinguish the wild-type target DNA from the mutant target DNA by one base mutation, performing successfully distinction for the CYP2C19 genotypes.
Figure 3. Specificity and sensitivity of the PCR-GoldMag LFA system. (a) The specificity results of LFA system presents. (b) Different DNA quantities of CYP2C19*2 and CYP2C19*3 heterozygous mutation genotype presents the sensitivity of LFA system. (c) Schematic of the sensitivity results of magnetic reader. (d) The magnetic signals of different DNA quantities of CYP2C19*2 and CYP2C19*3 heterozygous mutation genotype.
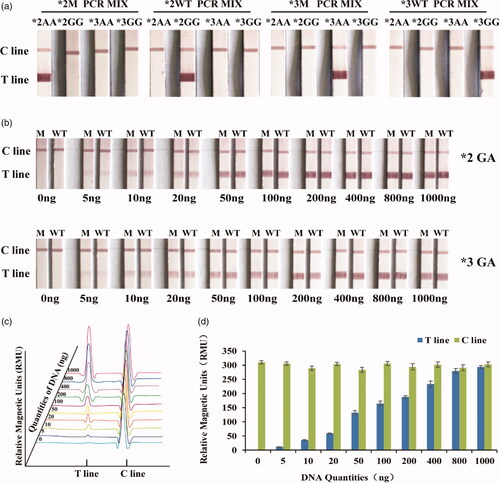
The PCR products amplified with different quantities (5, 10, 20, 50, 100, 200, 400, 800, and 1000 ng, and 0 ng as a negative control) of genomic DNA as templates were run through LFA system, respectively to test the sensitivity of this method. It presented the images of LFA system by different DNA quantities of CYP2C19*2 and CYP2C19*3 heterozygous mutation. The signal intensity correspondingly increased as the DNA quantities augmented. The red bands on the T line could be observed by visually () or by a magnetic reader () when PCR products amplified with greater or equal to 5 ng of genomic DNA, which was considered to be the limit of detection of this system. The magnetic signal value significantly improved as the DNA quantities increased ().
The reproducibility of the PCR-GoldMag LFA system was assessed by CYP2C19 genotypes, which were identified by DNA sequencing. Each genotype was run with the LFA system in triplicate with different batches of strips. The result indicated that there was no significant distinction among the three pair strips (Figure S7).
Under the optimized conditions, the PCR-GoldMag LFA system for CYP2C19 genotypes demonstrates good specificity. It is a crucial impact on the specificity of our method because the mismatches at the 3rd nucleotide from the 3′-ends of the primers were introduced [Citation27]. The limit of detection of PCR-LFA can reach 5 ng of the starting genomic DNA as a template. Technically, the sensitivity of the LFA system for clinical samples detection depends on the capacity of antibody conjugation upon nanoparticles. In our system, the conjugated antibodies can achieve more than 100 μg mg−1 but other kinds of nanoparticles with antibodies are difficult to reach 50 μg mg−1 due to the structure of GMNPs, which has large specific surface. This ensures the sensitivity of the LFA system [Citation17,Citation28].
Clinical application
To further validate the PCR-GoldMag LFA system for the CYP2C19 genotypes, 1356 samples from three hospitals were analyzed by this method. The DNA sequencing was used as a comparison. These samples satisfied the Hardy–Weinberg Law (CYP2C19*2, χ2 = 0.033, p = .984; CYP2C19*3, χ2 = 1.615, p = .446), which signified it was a reliable group representative. By chi-square test, there was no statistical difference between these two methods (χ2 = 0.002, p = 1 > .05). Kappa coefficient is 0.999 indicating that the detection results of the two methods have achieved a satisfactory degree of consistency. Among the 1356 samples, 1355 test results were consistent with the sequencing results and the total coincidence rate was 99.93%. There were 554 cases of CYP2C19*1/*1, with a coincidence rate of 99.82%. Other five genotypes were 100% (). Therefore, it is proved that the PCR-GoldMag LFA system is reliable for CYP2C19 genotyping.
Table 3. The coincidence rate of PCR-GoldMag LFA system and DNA sequencing.
In addition, we studied the distribution frequency of CYP2C19 genotypes in 1356 clinical samples. The proportion of *1/*1(555 cases) and *1/*2(494 cases) is the largest which is about 77.36%. *3/*3 (12 cases) is 0.88%. According to the metabolizer type, the extensive metabolizer (EM, *1/*1) accounted for 40.93%, the intermediate metabolizer type (IM, *1/*2, *1/*3) accounted for 43.22% and the poor metabolizer (PM, 2/*2, *2/*3, *3/*3) accounted for 15.85%. We also respectively analyzed the genotype distribution of the CYP2C19*2 and *3. The genotype distribution of *2 was GG 48.60%, GA 42.03%, AA 9.37% and the allele frequency was G 69.62%, A 30.38%. The genotype distribution of *3 was GG 86.73%, GA 12.39%, AA 0.88%, and the allele frequency was G 92.92%, A 7.08% ().
Figure 4. Pie chart for the genotype and allele frequency of CYP2C19 with 1356cases. (a) Gene frequency of CYP2C19. (b) Metabolic type of EM, IM, and PM. (c) The sample size data for genotyping of two provinces. (d) Genotype frequencies of CYP2C19*2 and CYP2C19*3.
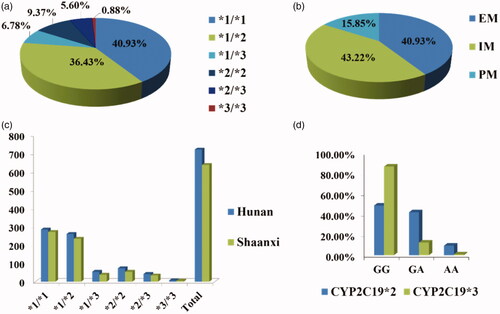
The allelic frequencies of CYP2C19 vary substantially around the world due to the differences of geographic and racial. Previous studies reported that the distribution of CYP2C19 alleles is slightly different in Chinese [Citation29–31]. Hu et al. [Citation31] showed that the distribution of CYP2C19 alleles in China, including 2127 clinical samples, with the distribution of *2 and *3 alleles at 33.07 and 5.34%. Our study showed that the allele frequency of CYP2C19*2 and CYP2C19 *3 are 30.38 and 7.08% in Han Chinese, respectively, which is similar to the study by Hu LM (n = 2127) with no significant difference in allele distribution (χ2 = 0.463, p = .496 > .05). CYP2C19*2 has the frequency significantly higher in Chinese Han populations than in others (15% in Caucasian and 19.2% in African) [Citation32]. The incidence of CYP2C19*3 is 0.04, 0.4, and 7.08% in Caucasians, African Americans [Citation14], and Han Chinese, respectively. Furthermore, we compared the distribution of CYP2C19 in Hunan and Shaanxi province and the data showed that there was no significant difference of CYP2C19 distribution between Hunan province (southern) and Shaanxi province (northern) populations (χ2 = 3.283, p = .657 > .05).
Conclusions
In this study, we established a PCR-GoldMag lateral flow assay system for CYP2C19 genotyping, using our patented gold magnetic nanoparticles as a carrier. Visual or automatic interpretation of genotyping results could be obtained in 5 min. The concordance rate between the LFA system and sequencing is 99.93%. Our data have demonstrated that this assay is highly useful for CYP2C19 genotyping in clinical practice due to its advantages, such as superior sensitivity and specificity, low cost, easy operation and no need for sophisticated instruments. This PCR-GoldMag LFA system provides a new way in the clinical application of CYP2C19 genotyping for guiding the use of clopidogrel. It also can be further adapted for other drug metabolism-related genes as well as SNPs in the field of personalized medicine.
Supplement_Materials.docx
Download ()Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Hou X, Han W, Gan Q, et al. CYP2C19 and ABCB1 genetic polymorphisms correlate with the recurrence of ischemic cardiovascular adverse events after clopidogrel treatment. J Clin Lab Anal. 2018;32:e22369.
- Matetzky S, Shenkman B, Guetta V, et al. Clopidogrel resistance is associated with increased risk of recurrent atherothrombotic events in patients with acute myocardial infarction. Circulation. 2004;109:3171–3175.
- Jia DM, Chen ZB, Zhang MJ, et al. CYP2C19 polymorphisms and antiplatelet effects of clopidogrel in acute ischemic stroke in China. Stroke. 2013;44:1717–1719.
- Jeong YH, Tantry US, Kim IS, et al. Effect of CYP2C19*2 and *3 loss-of-function alleles on platelet reactivity and adverse clinical events in East Asian acute myocardial infarction survivors treated with clopidogrel and aspirin. Circ Cardiovasc Interv. 2011;4:585–594.
- Grant JR, Arantes AS, Liao X, et al. In-depth annotation of SNPs arising from resequencing projects using NGS-SNP. Bioinformatics. 2011;27:2300–2301.
- Ahsan M, Li X, Lundberg AE, et al. Identification of candidate genes and mutations in QTL regions for chicken growth using bioinformatic analysis of NGS and SNP-chip data. Front Genet. 2013;4:226.
- Radvansky J, Resko P, Surovy M, et al. High-resolution melting analysis for genotyping of the myotonic dystrophy type 1 associated Alu insertion/deletion polymorphism. Anal Biochem. 2010;398:126–128.
- Harrington CT, Lin EI, Olson MT, et al. Fundamentals of pyrosequencing. Arch Pathol Lab Med. 2013;137:1296–1303.
- Howarth R, Yearwood C, Harvey JF. Application of dHPLC for mutation detection of the fibrillin-1 gene for the diagnosis of Marfan syndrome in a National Health Service Laboratory. Genet Test. 2007;11:146–152.
- Kriegsmann M, Arens N, Endris V, et al. Detection of KRAS, NRAS, and BRAF by mass spectrometry – a sensitive, reliable, fast and cost-effective technique. Diagn Pathol. 2015;10:132.
- Martinez-Serra J, Robles J, Nicolas A, et al. Fluorescence resonance energy transfer-based real-time polymerase chain reaction method without DNA extraction for the genotyping of F5, F2, F12, MTHFR, and HFE. J Blood Med. 2014;5:99–106.
- Loo KW, Griffiths LR, Gan SH. A novel multiplex PCR-RFLP method for simultaneous detection of the MTHFR 677 C > T, eNOS +894 G > T and – eNOS – 786 T > C variants among Malaysian Malays. BMC Med Genet. 2012;13:34.
- Roberts DG, Morrison TB, Liu-Cordero SN, et al. A nanoliter fluidic platform for large-scale single nucleotide polymorphism genotyping. BioTechniques. 2009;46:ix–xiii.
- Zhou SF, Liu JP, Chowbay B. Polymorphism of human cytochrome P450 enzymes and its clinical impact. Drug Metab Rev. 2009;41:89–295.
- Tost J, Gut IG. Genotyping single nucleotide polymorphisms by MALDI mass spectrometry in clinical applications. Clin Biochem. 2005;38:335–350.
- Yang D, Ma J, Zhang Q, et al. Polyelectrolyte-coated gold magnetic nanoparticles for immunoassay development: toward point of care diagnostics for syphilis screening. Anal Chem. 2013;85:6688–6695.
- Wenli HFS, Yan K, Peng M, et al. Fe3O4/Au/Fe3O4 nanoflowers exhibiting tunable saturation magnetization and enhanced bioconjugation. Nanoscale. 2012;4:747–751.
- Mieszawska AJ, Mulder WJ, Fayad ZA, et al. Multifunctional gold nanoparticles for diagnosis and therapy of disease. Mol Pharm. 2013;10:831–847.
- Bagheri S, Yasemi M, Safaie-Qamsari E, et al. Using gold nanoparticles in diagnosis and treatment of melanoma cancer. Artif Cells Nanomed Biotechnol.2018;46:462–471.
- Golchin K, Golchin J, Ghaderi S, et al. Gold nanoparticles applications: from artificial enzyme till drug delivery. Artif Cells Nanomed Biotechnol. 2018;46:250–254.
- Hui W, Zhang S, Zhang C, et al. A novel lateral flow assay based on GoldMag nanoparticles and its clinical applications for genotyping of MTHFR C677T polymorphisms. Nanoscale. 2016;8:3579–3587.
- Rastogi SK, Gibson CM, Branen JR, et al. DNA detection on lateral flow test strips: enhanced signal sensitivity using LNA-conjugated gold nanoparticles. Chem Commun. 2012;48:7714–7716.
- Newton CR, Graham A, Heptinstall LE, et al. Analysis of any point mutation in DNA. The amplification refractory mutation system (ARMS). Nucleic Acids Res. 1989;17:2503–2516.
- Little S. Amplification-refractory mutation system (ARMS) analysis of point mutations. Curr Protoc Hum Genet. 2001;Chapter 9:Unit 9.8. New York (NY): John Wiley & Sons, Inc; 2001. p. 9.8.1−9.8.12.
- Liu J, Huang S, Sun M, et al. An improved allele-specific PCR primer design method for SNP marker analysis and its application. Plant Methods. 2012;8:34.
- Cui Y, Hu D, Fang Y, et al. Preparation and mechanism of Fe3O4/Au core/shell super-paramagnetic microspheres. Sc China Ser B-Chem. 2001;44:404–410.
- Chen F, Zhao Y, Fan C, et al. Mismatch extension of DNA polymerases and high-accuracy single nucleotide polymorphism diagnostics by gold nanoparticle-improved isothermal amplification. Anal Chem. 2015;87:8718–8723.
- Yang D, Ma J, Xue C, et al. One-pot synthesis of poly (acrylic acid)-stabilized Fe3O4 nanocrystal clusters for the simultaneously qualitative and quantitative detection of biomarkers in lateral flow immunoassay. J Pharm Biomed Anal. 2018;159:119–126.
- Zhou Q, Yu XM, Lin HB, et al. Genetic polymorphism, linkage disequilibrium, haplotype structure and novel allele analysis of CYP2C19 and CYP2D6 in Han Chinese. Pharmacogenomics J. 2009;9:380–394.
- Chen L, Qin S, Xie J, et al. Genetic polymorphism analysis of CYP2C19 in Chinese Han populations from different geographic areas of mainland China. Pharmacogenomics. 2008;9:691–702.
- Hu LM, Dai DP, Hu GX, et al. Genetic polymorphisms and novel allelic variants of CYP2C19 in the Chinese Han population. Pharmacogenomics. 2012;13:1571–1581.
- Man M, Farmen M, Dumaual C, et al. Genetic variation in metabolizing enzyme and transporter genes: comprehensive assessment in 3 major East Asian subpopulations with comparison to Caucasians and Africans. J Clin Pharmacol. 2010;50:929–940.