Abstract
In this study, we identified hsa_circ_0051240 was significantly increased in ovarian cancer (OC) tissues. Our results indicated that silencing of hsa_circ_0051240 inhibited OC cell proliferation, migration and invasion in vitro, and also prevented OC tumour formation in vivo. In addition, the inhibitory effects by blockage of hsa_circ_0051240 could be attenuated by the miR-637 inhibitor. Furthermore, we also identified that hsa_circ_0051240 act as a sponge of miR-637, and miR-637 directly targeted KLK4 mRNA in OC cells. Altogether, hsa_circ_0051240 promotes OC cell proliferation, migration and invasion through inhibiting the miR-637/KLK4 axis. Therefore, these results demonstrated that the hsa_circ_0051240/miR-637/KLK4 axis might serve as a therapeutic target for OC treatment.
Introduction
It is reported that more than 70% ovarian cancer (OC) patients would experience recur followed by standard treatment [Citation1,Citation2]. These challenges make us urgently need to identify potential biomarkers for early diagnosis and design more targeted drugs for therapy. Circular RNA (circRNA) was discovered ubiquitous expressing in eukaryotic genes in 2012, and it constitutes the dominant isoform in hundreds of human genes [Citation3,Citation4]. Recently, it became a new focus in the research of ceRNA family [Citation5]. Functional circRNAs have been proved to act as miRNA sponges and RNA-binding protein sequestering agents as well as transcriptional regulators [Citation6]. In the present study, the hsa_circ_0051240 was an exon circRNA transcript from the NM_004363 fragment of chromosome 19 (chr19:42224841–42231272) and can be formed in exon 4 and exon 5 from the CEACAM5 gene. However, the function and mechanism of hsa_circ_0051240 in OC progress have not been reported.
As another kind of non-coding RNAs, microRNAs (miRNAs) can negatively regulate gene expression at the post-transcriptional level [Citation7]. Thus, hundreds of literature indicated miRNAs have significant effects on the regulation of tumour biology and inflammation [Citation8]. MicroRNA-637 (miR-637) was first identified by Cummins, J. M. and his colleagues from colorectal tumour tissues in 2006 [Citation9]. In addition, miR-637 has been found to be down-regulated and act as a tumour suppressor gene in various tumour types, including hepatocellular carcinoma (HCC) [Citation10], cholangiocarcinoma (CCA) [Citation11] and pancreatic ductal adenocarcinoma (PDAC) [Citation12]. Till now, several genes have been verified to be targeted by miR-637, such as Akt1, Osterix, and Cyclin-Dependent Kinase 6 (CDK6) [Citation13–15]. However, the function and mechanism of miR-637 in OC are not entirely clear.
In the present study, we firstly identified that hsa_circ_0051240 is significantly increased in OC tissues and explored the clinic significance of elevated hsa_circ_0051240 in OC patients. Then we investigated the roles of hsa_circ_0051240 on OC cell functions in vitro and in vivo. In addition, we explored the carcinogenic effects of hsa_circ_0051240 as a natural miR-637 sponge to modulate Kallikrein 4 (KLK4) expression in OC. Furthermore, we proved that hsa_circ_0051240 promoted cell proliferation, migration and invasion of OC by inhibition of miR-637 and activation of KLK4. Therefore, we demonstrated that the hsa_circ_0051240/miR-637/KLK4 axis plays critical roles in the OC pathogenesis and development, and might be expected to be a new therapy of OC.
Materials and methods
Patients and specimens
The pairs of OC and corresponding adjacent non-cancerous tissues were collected from patients who had received surgery in Harbin Medical University Cancer Hospital from December 2016 to July 2018. The essential clinicopathologic features of OC patients are shown in . All patients had not received preoperative treatment and the neoplasm staging was in accordance with the International Federation of Gynecology and Obstetrics (FIGO) Staging. Samples were stored at −80 °C rapidly for the next experiments. The study was sanctioned by the Ethics Committee of the Harbin Medical University Cancer Hospital. Written informed consent was obtained from the patients prior to participation.
Table 1. Correlation between relative has_circ_0051240 expression and clinicopathological characteristics of ovarian cancer patients (dCt).
Cell culture
Four OC cell lines (CAOV-3, SKOV-3, OVCAR-3 and H8910) and human ovarian epithelial (HOSE) cells were purchased from the Cell Bank of China Academy of Sciences, Shanghai, China. All the OC cells were cultured in Roswell Park Memorial Institute medium 1640 (Invitrogen, Carlsbad, CA, USA), while HOSE cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Invitrogen, Carlsbad, CA, USA). All the mediums contained with 10% foetal bovine serum (FBS; Gibco, Grand Island, NY, USA) and 100 U/ml penicillin–streptomycin. All cells were cultured in a humidified incubator at 37°C with 5% CO2 atmosphere.
Microarray analysis
Total RNA was isolated from 10 pairs of OC specimens, and the enriched circular RNAs were amplified after digesting with RNase R (Epicentre, Inc.). Then, the RNA was quantified by the NanoDrop ND-1000 (Thermo Scientific, Wilmington, DE, USA). Next, the circular RNA microarray hybridization was performed using Arraystar Super RNA Labeling Kit (Arraystar Super RNA Labeling Kit; Arraystar) according to the manufacturer's protocols. Afterwards, the labeled cRNAs were hybridized onto the Arraystar Human circRNA Array (8 × 15 K, Arraystar). And the arrays were then scanned by the Agilent Scanner G2505C.
Oligonucleotide transfection
The small interfering RNAs (siRNA) and negative control (NC) oligonucleotides utilized for cell transfection were synthesized by GenePharma (Shanghai, China). H8910 and OVCAR-3 cells were grown in 6-well plates and were transiently transfected with si-hsa_circ_0051240 or NC using Lipofectamine 2000 (Invitrogen, CA, USA) following the manufacturer’s protocol.
RNA extraction and quantitative real-time PCR (qRT-PCR) assay
The RNA was extracted from specimens and cells by using TRIzol (Invitrogen; Thermo Fisher Scientifc, Inc., Waltham, MA, USA). cDNAs were synthesized by Moloney Murine Leukemia Virus reverse transcriptase (Promega Corporation, Madison, WI, USA) and oligo dT 15 primers (Thermo Fisher Scientific, Inc.). Primers were designed to perform the amplification, and the sequences were as follows: GAPDH, Forward: 5′-TATGATGATATCAAGAGGGTAGT-3′ and Reverse: 5′-TGTATCCAAACTCATTGTCATAC-3′; U6, Forward: 5′-CTCGCTTCGGCAGCACATA-3′ and Reverse:5′-AACGATTCACGAATTTGCGT-3′; hsa_circ_0051240, Forward: 5′-TGACGGACGATTCAGCATCT-3′ and Reverse: 5′-ATACTGCGGGGATGGGTTAG-3′; miR-637, Forward: 5′- ACTGGGGGCTTTCGGG-3′ and Reverse: 5′-AGTGCGTGTCGTGGAGTC-3′.
KLK4 Forward: 5′-AGCTCTATGACCCGCTGTAC-3′ and Reverse: 5′- TGCAGAGGTTGGTGTAGACA-3′. The amplification of target genes was performed using the SYBR Select Master Mix (Applied Biosystems) then analyzed with the ABI7300 System (Applied Biosystems). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and U6 small nuclear RNA levels were used to normalize the expressions of hsa_circ_0051240, KLK4 and miR-637, respectively. Relative expressions were determined based on 2−ΔΔCt method [Citation16].
Dual-luciferase reporter assay
Hsa_circRNA_0051240-WT and hsa_circ_0051240-Mut, KLK4-WT and KLK4-Mut were inserted into pGL3-Basic vector (Promega, Madison, WI, USA) in Dual-luciferase reporter assay. Cells were plated in a 96-well plate and transfected with indicated reporter plasmid by applying Lipofectamine 2000 (Life Technologies). Then a Dual-luciferase Reporter assay system (Promega, Madison, WI, USA) was used to measure the luciferase activity. All experiments were repeated three times independently.
Pull-down assay
The pull-down assay with biotinylated miRNA was performed to determine the relationship between hsa_circ_0051240 and miR-637. In brief, OC cell lines H8910 and OVCAR-3 cells were transfected with biotinylated miR-637 mimics or mutant miR-637 using Lipofectamine RNAiMax (Life Technologies). After 48 h, the cells were sonicated and 50 μl cell lysates were aliquot for input. While the remaining cell lysates were incubated with C-1 magnetic beads (Life Technologies) at 4 °C for 3 h. RNeasy Mini Kit (QIAGEN, Germany) was used to purify the bound RNAs for the analysis.
CCK-8 assay
The proliferation ability of OC cells was determined using Cell Counting Kit-8 (CCK-8) (Dojindo Co. Ltd., Kumamoto, Japan). Briefly, following the manufacturer’s instructions, 10 µL of CCK-8 solution was added to each well (5000 cells/100 μl) of 96-well plates and incubated for 4 h at 37 °C. SpectraMax M5 ELISA plate reader (Molecular Devices, LLC, Sunnyvale, CA, USA) was used to measure the optical density value at 450 nm.
Colony formation assay
Briefly, 500 cells/well were plated into 6-well plates evenly. After incubated at 37 °C for 14 days, colonies were fixed by 4% paraformaldehyde and stained with 0.5% crystal violet. Next, the number of colonies containing at least 50 cells (established by microscopy) were counted. All experiments were repeated ≥3 times.
Migration and invasion assays
Cell migration and invasion assays were carried out with Transwell cell culture chambers (Corning Costar Corp, Cambridge, MA, USA) according to the manufacturer’s protocol. Briefly, a total of 5 × 105 cells (200 μl/well) resuspended in serum-free medium were seeded into the upper chamber, while the lower chamber was filled with 600 µL complete medium supplemented with 20% FBS. After incubation at 37 °C for 24 h, cells remained on the upper surface of the membrane were cleaned with cotton swabs, while cells passed through the filter were fixed by 4% paraformaldehyde and stained with 0.1% crystal violet solution. Then the migrated cells were photographed using a light microscope at 20 × objectives. For cell invasion assays, the membrane was pre-coated with diluted Matrigel, and the rest processes were executed as described above.
Xenografts in mice
A total of ten 6-week-old male nude mice were purchased from Shanghai LAC Laboratory Animal Co. Ltd. Control (negative cells). All animal studies were approved by Harbin Medical University Cancer Hospital Ethics Committee. 1 × 105 cells resuspended in physiological saline were injected into the back flank of mice. Then all the mice were housed in a specific‑pathogen‑free (SPF) grade animal house with 12 h light/dark cycle and constant temperature (25 °C). When a tumour was palpable (a week later), tumour sizes were then recorded every week with a caliper following the formula: V = L × W2 × 0.5 (L, length; W, width). After 5 weeks, the mice were executed by euthanasia, and the tumours were dissected and weighed.
Statistical analysis
SPSS (version 15.0 (SPSS, Inc., Chicago, IL, USA) and GraphPad Prism 6 (GraphPad Software, Inc., La Jolla, CA, USA) were performed for statistical analysis. The student’s t-test or one-way analysis of variance followed by a post hoc Tukey test was used for calculation of statistic differences. p < .05 was considered to be statistically significant.
Results
Identification of hsa_circ_0051240 in OC
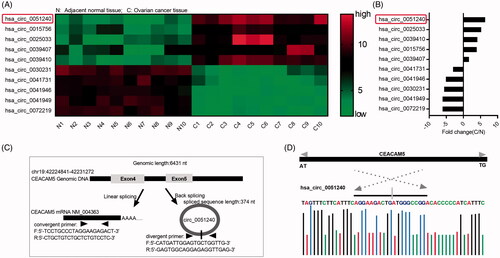
To explore circRNAs expression profile in OC tissues, we detected 10 pairs of OC tissue samples (10 adjacent normal tissues and 10 OC tissues) by CircRNA microarray. According to the expression intensity of circRNAs, we screened Top 5 upregulated and Top 5 downregulated circRNAs (). The fold changes (OC tissue/adjacent normal tissue, C/N) of Top 5 upregulated and Top 5 downregulated circRNAs were also shown (). Among them, we found that hsa_circ_0051240 expression was consistently and significantly increased in OC tissues compared with the matched adjacent normal tissues (). As shown in , the hsa_circ_0051240 was an exon circRNA transcript from the NM_004363 fragment of chromosome 19 (chr19:42224841–42231272). Hsa_circ_0051240 can be formed in exon 4 and exon 5 from the CEACAM5 gene. CEACAM5 mRNA can be amplified by using convergent primers, and hsa_circ_0051240 can be amplified by using divergent primers. In addition, the distinct product of hsa_circ_0051240 was confirmed by Sanger sequencing ().
Figure 1. Identification of hsa_circ_0051240 in OC. (A) The cluster heat map of Top 5 upregulated and Top 5 downregulated circRNAs in adjacent normal tissues (n = 10) and OC tissues (n = 10). Green represents low expression; red represents high expression. Individual tissue is described as a column. (B) The fold changes (C/N) of Top 5 upregulated and Top 5 downregulated circRNAs were shown. C: OC tissue; N: adjacent normal tissue. (C) The schematic diagram alternative splicing isoforms. Convergent primers detected CEACAM5 mRNA, divergent primers detected hsa_circ_0051240. (D) Sanger sequencing verified head-to-tail splicing.
High expression of hsa_circ_0051240 in OC
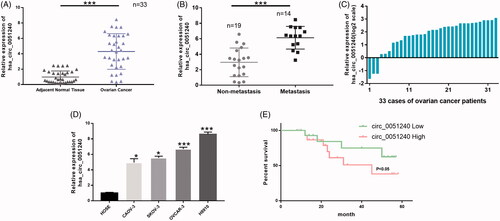
To further demonstrate hsa_circ_0051240 expression in OC, the qRT-PCR assay was used to analyze hsa_circ_0051240 expression in OC tissues and OC cell lines. Firstly, the results indicated that hsa_circ_0051240 expression was significantly increased in OC tissues compared with adjacent normal tissues (p < .001, ). We also found that hsa_circ_00512401 expression was significantly upregulated in OC patients with metastasis (n = 14) as compared to that in OC patients without metastasis (n = 19) (p < .001, ). The relative expression level of hsa_circ_0051240 was also shown in 33 cases of OC patients (log2 scale) (). The relative expression level of hsa_circ_0051240 was correlated with lymph node metastasis (p = .0031), the FIGO stage (p = .0008) and the CA125 level of patients (p = .0071), but not with age and ascites volume (). Secondly, the results revealed that hsa_circ_0051240 expression was significantly increased in OC cells (CAOV-3, SKOV-3, OVCAR-3 and H8910) relative to human ovarian epithelial (HOSE) cells (p < .05, p < .001, ). We also found that hsa_circ_0051240 expression was lower in OVCAR-3 cells than other OC cell lines, and higher in H8910 cells than other OC cell lines, we chose OVCAR-3 and H8910 cells as the research objectives. As hsa_circ_0051240 expression is significantly increased in OC, we next analyzed whether hsa_circ_0051240 expression is correlated with the prognosis of OC. A cohort of 33 OC patients with survival data showed that there was shorter survival time in OC patients with high expression of hsa_circ_0051240 than OC patients with low expression of has_circ_0051240, suggesting that increased hsa_circ_0051240 was significantly correlated with poor prognosis of OC patients (p < .05, ).
Figure 2. High expression of hsa_circ_0051240 in OC. (A) Relative expression level of hsa_circ_0051240 was accessed by qRT-PCR assay in adjacent normal tissues (n = 33) and OC tissues (n = 33) (***p < .001). (B) Hsa_circ_0051240 expression was measured by qRT-PCR assay in non-metastasis (n = 19) and metastasis (n = 14) (***p < .001). (C) Relative expression level of hsa_circ_0051240 in 33 cases of OC patients (log2 scale). (D) Hsa_circ_0051240 expression was analyzed by qRT-PCR assay in human ovarian epithelial (HOSE) cells and OC cells (CAOV-3, SKOV-3, OVCAR-3 and H8910) (*p < .05, ***p < .001). (E) Based on hsa_circ_0051240 expression, the overall survival of OC patients was analyzed by Kaplan-Meier analysis (p < .05).
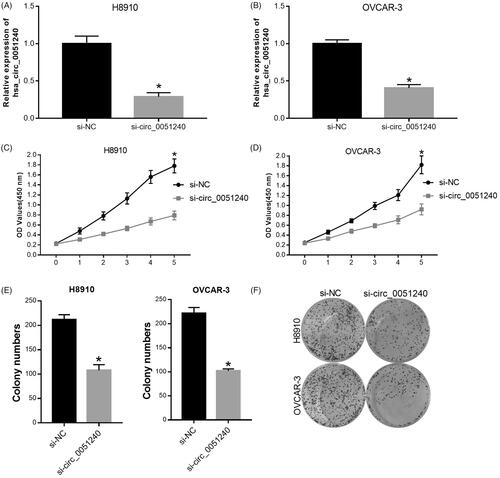
Silence of hsa_circ_0051240 inhibits OC cell proliferation
According to the above observations, we then demonstrated the function and mechanism of hsa_circ_0051240 in OC. Firstly, H8910 and OVCAR-3 cells were transfected with negative control (si-NC) and hsa_circ_0051240 siRNAs (si-circ_0051240), respectively. Hsa_circ_0051240 expression was analyzed in treated OVCAR-3 and H8910 cells, and the results testified that hsa_circ_0051240 was lowly expressed in si-circ_0051240 group compared with si-NC group (p < .05, ). In addition, the results from CCK-8 and colony formation assays revealed that silence of hsa_circ_0051240 significantly inhibited the proliferation abilities of OVCAR-3 and H8910 cells (p < .05, ).
Figure 3. Silence of hsa_circ_0051240 inhibits OC cell proliferation. H8910 and OVCAR-3 cells were transfected with negative control (si-NC) and hsa_circ_0051240 siRNAs (si-circ_0051240), respectively. (A, B) qRT-PCR assay was performed to evaluate hsa_circ_0051240 expression (*p < .05). (C, D) CCK-8 assay was performed to measure the proliferation abilities of H8910 and OVCAR-3 cells (*p < .05). (E, F) Clone formation assay was used to detect the colony-formation abilities of H8910 and OVCAR-3 cells (*p < .05).
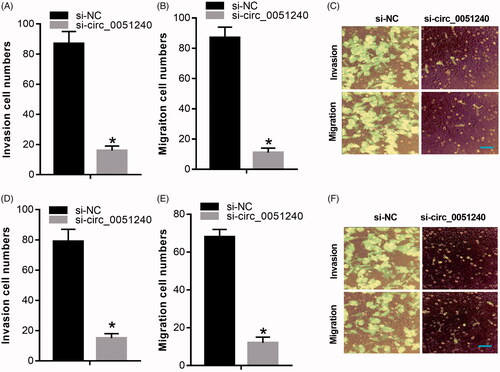
Silence of hsa_circ_0051240 suppresses OC cell migration and invasion
Base on the above, silence of hsa_circ_0051240 significantly inhibited OC cell proliferation, we further certified the effects of hsa_circ_0051240 on OC cell migration and invasion, and the data manifested that silence of hsa_circ_0051240 markedly inhibited the migration capacities of OVCAR-3 and H8910 cells (p < .05, ); silence of hsa_circ_0051240 also significantly suppressed the invasion abilities of OVCAR-3 and H8910 cells (p < .05, ).
Figure 4. Silence of hsa_circ_0051240 suppresses OC cell migration and invasion. H8910 and OVCAR-3 cells were transfected with si-NC and si-circ_0051240, respectively. (A–C) The migration capacity was tested by Transwell assay (*p < .05). (D–F) The invasion capacity was measured by Transwell assay (*p < .05).
Hsa_circ_0051240 negatively regulates miR-637
Research has proved that circRNAs can be used as a new kind of biomarker in various diseases by sponging multiple miRNAs [Citation17]. In our study, a total of 16 miRNAs that predicted interacted with has_circ_0051240 were obtained from circular RNA interactome, among which miR-637 had the highest context and score percentile (). Furthermore, we found that there was a binding site between miR-637 and hsa_circ_0051240 (), and then we detected the luciferase intensity of hsa_circ_0051240 by using the Dual-Luciferase Reporter Assay. As our results showed, miR-637 significantly decreased the luciferase intensity of wild-type hsa_circ_0051240, while miR-637 had no effect on the mutational hsa_circ_0051240 in H8910 and OVCAR-3 cells (*p < .05, ). Besides, our results indicated that silence of hsa_circ_0051240 significantly upregulated miR-637 expression in OC cells (*p < .05, ). Furthermore, the binding of hsa_circ_0051240 and miR-637 was measured by pull-down assay, and the results demonstrated that the relative fold enrichment of hsa_circ_0051240 was significantly increased in the miR-637-captured fraction group compared with NC group (p < .001, ).
Figure 5. Hsa_circ_0051240 negatively regulates miR-637. (A) The binding sites of miR-637 and the wild-type and mutated hsa_circ_0051240 were shown. (B, C) H8910 and OVCAR-3 cells were co-transfected with miR-637 or scramble, and wild-type or mutated hsa_circ_0051240, respectively. The luciferase intensity was measured by the Dual-Luciferase Reporter Assay (*p < .05). (D) miR-637 expression was estimated by qRT-PCR assay in H8910 and OVCAR-3 cells transfected with si-NC and si-circ_0051240 (*p < .05). (E, F) H8910 and OVCAR-3 cells were treated with bio-scramble, bio- miR-637, and mutant bio-miR-637, and binding of hsa_circ_0051240 and miR-637 was accessed by pull-down assay (***p < .001).
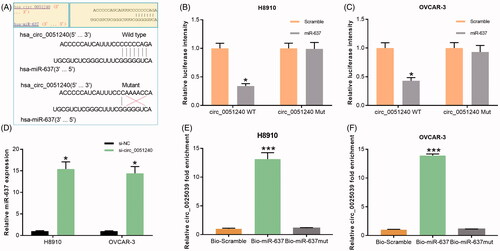
Table 2. miRNAs That predicted interacted with has_circ_0051240 by circular RNA interactome.
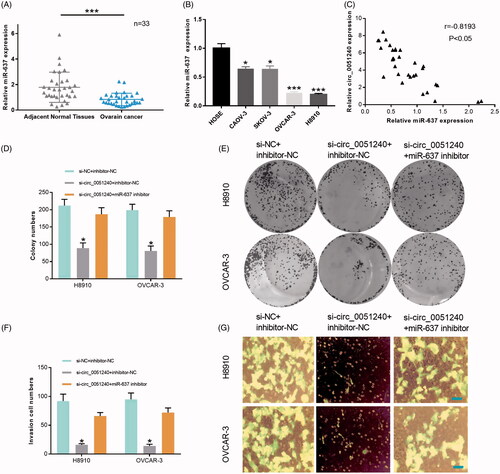
Silence of hsa_circ_0051240 inhibits OC cell proliferation, migration and invasion by miR-637
In our study, we also found that miR-637 expression was significantly decreased in OC tissues compared with adjacent normal tissues (p < .001, n = 33, ). MiR-637 expression was also significantly reduced in OC cells (CAOV-3, SKOV-3, OVCAR-3 and H8910) relative to HOSE cells (p < .05, p < .001, ). Therefore, miR-637 was lowly expressed in OC. In addition, we found that there was a negative correlation between hsa_circ_0051240 and miR-637 expression in OC tissues (r = −0.8193, p < .05, ). Functional experiments also indicated that silence of hsa_circ_0051240 significantly inhibited the proliferation abilities of the transfected H8910 and OVCAR-3 cells, and miR-637 inhibitor then attenuated the inhibitory effects of cell proliferation mediated by hsa_circ_0051240 silencing (p < .05, ). Similarly, the results also proved that silence of hsa_circ_0051240 significantly suppressed the invasion abilities of the transfected H8910 and OVCAR-3 cells, and miR-637 inhibitor then weakened the inhibitory effects of cell invasion mediated by hsa_circ_0051240 silencing (p < .05, ).
Figure 6. Silence of hsa_circ_0051240 inhibits OC cell proliferation, migration, and invasion by miR-637. (A) miR-637 expression was evaluated by qRT-PCR assay in adjacent normal tissues (n = 33) and OC tissues (n = 33) (***p < .001). (B) miR-637 expression was analyzed by qRT-PCR assay in HOSE, CAOV-3, SKOV-3, OVCAR-3 and H8910 cells (*p < .05, ***p < .001). (C) The correlation coefficient between hsa_circ_0051240 and miR-637 expression was analyzed by Pearson’s correlation algorithm (r = −0.8193, p < .05). (D, E) The colony-formation abilities were detected by colony formation assay in H8910 and OVCAR-3 cells transfected with si-circ_0051240 or miR-637 inhibitor as indicated (*p < .05). (F, G) the migration and invasion abilities were detected by Transwell assay in H8910 and OVCAR-3 cells transfected with si-circ_0051240 or miR-637 inhibitor as indicated (*p < .05).
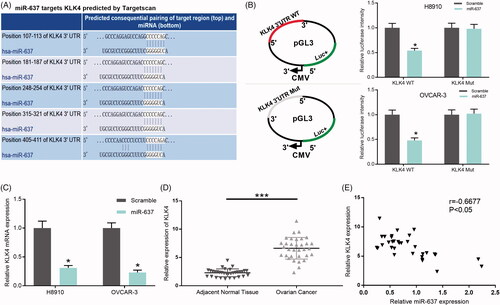
miR-637 negatively regulates KLK4
According to TargetScan prediction, we found that miR-637 targets Kallikrein-related peptidases (KLK4), and there were five putative complementary sites between KLK4 and miR-637 (). We then constructed the wild-type KLK4 3’UTR and mutant KLK4 3’UTR, H8910 and OVCAR-3 cells were co-transfected with miR-637 or scramble, and wild-type or mutated KLK4, respectively. The results from the Dual-Luciferase Reporter Assay indicated that miR-637 significantly decreased the luciferase intensity in wild-type KLK4, however, miR-637 cannot change the luciferase intensity of mutant KLK4 group (p < .05, ). In addition, we found that miR-637 significantly downregulated KLK4 expression in H8910 and OVCAR-3 cells (p < .05, ). In OC patients, we found that KLK4 expression was significantly increased in OC tissues compared with adjacent normal tissues (p < .001, n = 33, ), and there was a negative correlation between miR-637 and KLK4 expression in OC tissues (r = −0.6677, p < .05, ).
Figure 7. miR-637 negatively regulates KLK4. (A) The binding sites of miR-637 and the wild-type and mutated KLK4 were shown. (B) H8910 and OVCAR-3 cells were co-transfected with miR-637 or scramble, and wild-type or mutated KLK4, respectively. The luciferase intensity was measured by the Dual-Luciferase Reporter Assay (*p < .05). (C) KLK4 expression was measured by qRT-PCR assay in H8910 and OVCAR-3 cells transfected with scramble and miR-637 (*p < .05). (D) KLK4 expression was tested by qRT-PCR assay in adjacent normal tissues (n = 33) and OC tissues (n = 33) (***p < .001). (E) Pearson’s correlation algorithm was used to analyze the correlation coefficient between the expression of miR-637 and KLK4 (r = −0.6677, p < .05).
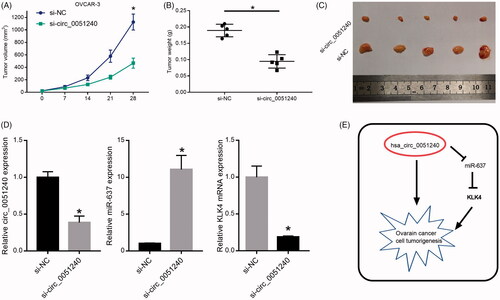
Hsa_circ_0051240 knock-down inhibits OC tumour formation in vivo
To further explored the effects of hsa_circ_0051240 on OC tumour formation in vivo, OVCAR-3 cells were transfected with si-NC and si-circ_0051240, respectively. Then, the nude mice were injected with the transfected cells (2 × 106 cells) and monitor at 7, 14, 21 and 28 days post-injection, respectively. The tumour volumes (mm3) and tumour weights (g) were counted and measured, and the results demonstrated that silence of hsa_circ_0051240 significantly decreased OC tumour volumes and weights (p < .05, ). The dissected tumours were also obtained and shown in . Moreover, our results indicated that silence of hsa_circ_0051240 significantly downregulated hsa_circ_0051240 and KLK4 expressions, and upregulated miR-637 expression (p < .05, ). Therefore, we suggested that hsa_circ_0051240 negatively regulates miR-637 by functioning as a miR-637 sponge, and miR-637 directly suppresses the promoter activity of KLK4, and downregulates KLK4 expression, which promotes OC cell tumourigenesis ().
Figure 8. Hsa_circ_0051240 knock-down inhibits OC tumour formation in vivo. OVCAR-3 cells were transfected with si-NC and si-circ_0051240, respectively. The nude mice were injected with the transfected cells (2 × 106 cells) for 7, 14, 21 and 28 days, respectively. (A) The parameters of tumour growth were measured, and the tumour volumes (mm3) were counted (*p < .05). (B) The tumour weights (g) were measured (*p < .05). (C) The dissected tumours were photographed, respectively. (D) Total RNA was extracted from the dissected tumours, and circ_0051240, miR-637 and KLK4 expressions were detected by qRT-PCR assay (*p < .05). (E) The diagrammatic sketch of the circ_0051240/miR-637/KLK4 axis in OC is shown.
Discussion
As a new type of endogenous noncoding RNAs, circRNAs have been recently identified play a major role in biological processes such as tumour cell proliferation, apoptosis, migration and invasion [Citation5]. Exploring the differentially expressed circRNAs and their functional mechanism in cancer progression has become a new research focus. In the present study, we first identified hsa_circ_0051240, derived from exon 4 and exon 5 of the CEACAM5 gene, was consistently and significantly increased in OC tissues. And the expression level of hsa_circ_0051240 was correlated with the poor prognosis of OC patients. Functionary, we proved hsa_circ_00512401 was significantly upregulated in diversified OC cells and silencing of hsa_circ_00512401 could inhibit OC cells proliferation, colony formation as well as migration and invasion abilities.
It’s reported that the competitive endogenous RNA (ceRNA) can affect the regulatory functions of miRNAs in gene expression and reverse the effect of miRNAs on certain pathological processes through a miRNA response element (MRE) [Citation18–20]. RNA transcripts with the same MREs can be competitive in inhibiting the function of miRNAs, thus acting as an RNA sponge to block the binding between miRNAs and their target genes [Citation5,Citation21]. Recent studies have shown that exonic circRNAs can function as ceRNA to regulate the expression of genes by adsorbing miRNAs, thus sponging for miRNAs [Citation18,Citation22,Citation23]. And miRNA sponge effects achieved by circRNA formation are now regarded as a general phenomenon since ciRS-7 (circRNA sponge for miR-7) and sex-determining region Y (Sry, testis-specific circRNA sponge for miR-138) had been discovered by Thomas B. Hansen and his colleagues in 2013 [Citation22]. A circRNA could have multiple miRNA binding sites. For instance, circHIPK3, elevated in various cancer types, serves as a sponge for 9 miRNAs (miR-124, miR-152, miR-193a, miR-29a, miR-29b, miR-338, miR-379, miR-584 and miR-654) [Citation24]. Besides, another circRNA, circ_001569 is reported to been upregulated in colorectal cancer (CRC) tissues and promotes CRC cell progression by sponging miR-145 [Citation25]. In the present study, we first identified that miR-637, a famous tumour suppressor gene, is a hsa_circ_0051240 associated miRNA. The fact that inhibition of hsa_circ_0051240 could up-regulate the expression level of miR-637 in OC cells, while miR-637 inhibitor could attenuate the anti-tumour effects mediated by hsa_circ_0051240 inhibition, suggesting that silencing of hsa_circ_0051240 inhibits OC progression though acting as miR-637 sponge.
Researches on human KLK gene family has been going on for decades, and great advances have been achieved in the understanding of the tissue and cellular distribution and physiological (and pathophysiological) functions of most KLKs [Citation26]. The prognostic significance of several KLKs in cancers has been well documented. For instance, KLK3, also called PSA, is now widely used for a tumour marker of prostate cancer (PC) and have been associated with prognosis and mortality [Citation27]. Likewise, it had been reported that elevated expression of KLK 4, is strongly associated with the poor prognosis in CRC and OC [Citation28,Citation29]. In our study, we first identified KLK4 as one of the target genes of miR-637 and confirmed it by using the Dual-Luciferase Reporter Assay. In addition, we found KLK4 expression was significantly increased in OC tissues and negatively correlated with miR-637 expression in OC tissues. Thus, miR-637 can directly target KLK4 and decrease its mRNA expression in OC.
Conclusion
Our study proved that hsa_circ_0051240 is up-regulated in OC and correlated with the lymph node metastasis and the FIGO stage. In addition, we also confirmed that hsa_circ_0051240 can promote OC cell growth and proliferation, migration and invasion through miR-637/KLK4 axis. Therefore, we suggested that hsa_circ_0051240 may act as a potential biomarker for OC patients, and provide a novel insight that inhibition of hsa_circ_0051240 might be a potential therapeutic strategy for OC.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Zhang T. Core signaling pathways in ovarian cancer stem cell revealed by integrative analysis of multi-marker genomics data. PLoS One. 2018;13:e0196351.
- Hennessy BT, Coleman RL, Markman M. Ovarian cancer. Lancet. 2009;374:1371–1382.
- Salzman J. Circular RNA expression: its potential regulation and function. Trends Genet. 2016;32:309–316.
- Salzman J, Gawad C, Wang PL, et al. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7:e30733.
- Zhong Y, Du Y, Yang X, et al. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol Cancer. 2018;17:79.
- Ebbesen KK, Kjems J, Hansen TB. Circular RNAs: identification, biogenesis and function. Biochim Biophys Acta. 2016;1859:163–168.
- Chatterjee N, Rana S, Espinosa-Diez C, et al. MicroRNAs in cancer: challenges and opportunities in early detection, disease monitoring, and therapeutic agents. Curr Pathobiol Rep. 2017;5:35–42.
- Vannini I, Fanini F, Fabbri M. Emerging roles of microRNAs in cancer. Curr Opin Genet Dev. 2018;48:128–133.
- Cummins JM, He Y, Leary RJ, et al. The colorectal microRNAome. Proc Natl Acad Sci USA. 2006;103:3687–3692.
- Zhang J-f, He M-l, Fu W-m, et al. Primate-specific microRNA-637 inhibits tumourigenesis in hepatocellular carcinoma by disrupting signal transducer and activator of transcription 3 signaling. Hepatology. 2011;54:2137–2148.
- Li JX, Ding XM, Han S, et al. Mir-637 inhibits the proliferation of cholangiocarcinoma cell QBC939 through interfering CTSB expression. Eur Rev Med Pharmacol Sci. 2018;22:1265–1276.
- Xu RL, He W, Tang J, et al. Primate-specific miRNA-637 inhibited tumourigenesis in human pancreatic ductal adenocarcinoma cells by suppressing Akt1 expression. Exp Cell Res. 2018;363:310–314.
- Zhang J-f, Fu W-m, He M-l, et al. MiR-637 maintains the balance between adipocytes and osteoblasts by directly targeting Osterix. Mol Biol Cell. 2011;22:3955–3961.
- Sang HY, Jin YL, Zhang WQ, et al. Downregulation of microRNA-637 increases risk of hypoxia-induced pulmonary hypertension by modulating expression of cyclin dependent kinase 6 (CDK6) in pulmonary smooth muscle cells. Med Sci Monit. 2016;22:4066–4072.
- Que T, Song Y, Liu Z, et al. Decreased miRNA-637 is an unfavorable prognosis marker and promotes glioma cell growth, migration and invasion via direct targeting Akt1. Oncogene. 2015;34:4952–4963.
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408.
- Kulcheski FR, Christoff AP, Margis R. Circular RNAs are miRNA sponges and can be used as a new class of biomarker. J Biotechnol. 2016;238:42–51.
- He J, Xie Q, Xu H, et al. Circular RNAs and cancer. Cancer Lett. 2017;396:138–144.
- Poliseno L, Salmena L, Zhang J, et al. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–1038.
- Cesana M, Cacchiarelli D, Legnini I, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–369.
- Salmena L, Poliseno L, Tay Y, et al. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358.
- Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388.
- Bezzi M, Guarnerio J, Pandolfi PP. A circular twist on microRNA regulation. Cell Res. 2017;27:1401–1402.
- Zheng Q, Bao C, Guo W, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215.
- Xie H, Ren X, Xin S, et al. Emerging roles of circRNA_001569 targeting miR-145 in the proliferation and invasion of colorectal cancer. Oncotarget. 2016;7:26680–26691.
- Prassas I, Eissa A, Poda G, et al. Unleashing the therapeutic potential of human kallikrein-related serine proteases. Nat Rev Drug Discov. 2015;14:183–202.
- Hannu K, Johanna M, Ulf-Håkan S. KLK-targeted therapies for prostate cancer. Ejifcc. 2014;25:207–218.
- Kontos CK, Chantzis D, Papadopoulos IN, et al. Kallikrein-related peptidase 4 (KLK4) mRNA predicts short-term relapse in colorectal adenocarcinoma patients. Cancer Lett. 2013;330:106–112.
- Shahinian H, Loessner D, Biniossek ML, et al. Secretome and degradome profiling shows that Kallikrein-related peptidases 4, 5, 6, and 7 induce TGFbeta-1 signaling in ovarian cancer cells. Mol Oncol. 2014;8:68–82.
