Abstract
This study aimed to explore the effect of cell division cycle protein 42 (CDC42) on inflammatory response and immune response in mice bearing inflammatory bowel disease (IBD). Trinitrobenzene sulfonic acid was injected into the colon of mice to establish IBD model. The mice were divided into four groups (n = 4): control, model, Ad5, and Ad5-CDC42. After establishing IBD model, mice which were treated with AD5 empty vector and AD5-CDC42 expression vector served as the Ad5 group and Ad5-CDC42 group, respectively. The mRNA and protein levels of interleukin 10 (IL-10), interferon-γ (IFN-γ), IL-4, and tumor necrosis factor-α (TNF-α) in the colon tissues were evaluated by RT-PCR and western blot, respectively. Their levels in the serum and colon tissues were examined by ELISA assay and immunohistochemical analysis, respectively. Their changes in the mRNA and protein levels were consistent and similar changes in the colon tissues and the serum were found among various groups. The levels of IL-10, IFN-γ, IL-4, and TNF-α were lowest in the control group. Their levels in the model group and the Ad5 group were similar (p > .05) and significantly higher than those in the control group (p < .05). In comparison with the model group and the Ad5 group, their levels were significantly reduced in the Ad5-CDC42 group (p < .05). In conclusion, the levels of inflammatory cytokines were elevated in the colon tissues and serum of IBD mice, which could be reduced by the CDC42 treatment. CDC42 regulated the inflammatory response and the innate immune response in IBD mice.
Introduction
Intestinal mucosal damage causes pathogenic microorganisms to invade the body, triggering a strong immune response and eventually leading to inflammatory bowel disease (IBD) [Citation1,Citation2]. A large number of migration, phagocytosis, development, and transformation of immune cells and macrophages are involved in immune responses [Citation3]. Macrophages not only have the ability to phagocytose pathogens, but also can secrete pro-inflammatory cytokines and anti-inflammatory cytokines, thereby regulating inflammation and playing a central role in the immune response against pathogenic microorganisms.
Cell division cycle protein 42 (CDC42) is a small-molecule switch protein, whose main function is to affect cell migration and polarity by regulating actin rearrangement [Citation4]. CDC42 can regulate the differentiation of immune cells [Citation5]. Moreover, CDC42 has great influence on macrophages. It participates in the regulation of the phagocytosis and migration of macrophage [Citation5,Citation6]. It binds to target proteins: (i) such as nonkinases-Wiskott-Aldrich syndrome proteins (WASPs), partitioning-defective protein 6 (PAR6), and IQ-motif-containing GTPase-activating proteins (IQGAPs) to regulate vesicle trafficking, cytoskeletal reorganization, and cell polarization; (ii) such as lipid kinase-phosphatidylinositol 3-kinase (PI3K) to regulate cell mitosis; (iii) such as serine/threonine kinases-p21-activated kinase (PAK), S6 kinase (S6K), mixed lineage kinase 3 (MLK3) to regulate transcriptional activity, cell cycle, and cytoskeletal reorganization; (iv) such as tyrosine kinase-acetate kinase (ACK) to regulate receptor-mediated endocytosis [Citation7–9].
Therefore, the relationship between CDC42 and the inflammatory response is an attractive research subject in IBD research, which should be explored. This study aimed to explore the effect of CDC42 on colonic inflammatory response and immune response in mice bearing IBD. Colonic inflammation was induced by injecting trinitrobenzene sulfonic acid (TNBS) into the colon of mice [Citation10]. Two proinflammatory cytokines (tumor necrosis factor-α (TNF-α) and interferon-γ (IFN-γ)) and two anti-inflammatory cytokines (interleukin 4 (IL-4) and IL-10) were selected as indicators to characterize the changes in the innate immune response and inflammatory response of IBD mice following CDC-42 treatment.
Materials and methods
Materials and animals
Adenovirus vectors were purchased from Shanghai Novobio Biotechnology Co., Ltd. (Shanghai, China). Lysis buffer (C1053) was procured from Applygen (Beijing, China). Mouse antiglyceraldehyde phosphate dehydrogenase (GAPDH) monoclonal antibody (TA-08), herseradish peroxidase (HRP)-conjugated goat anti-mouse IgG(H + L) (ZB-2305) and HRP-conjugated goat antirabbit IgG(H + L) (ZB-2301) were bought from ZSGB-Bio (Beijing, China). Mouse IL-4 ELISA kit (ml002149), mouse IL-10 ELISA kit (ml037873), mouse TNF-α ELISA kit (ml002095). and mouse IFN-γ ELISA kit (ml002277) were purchased from Shanghai Enzyme-linked Biotechnology Co., Ltd. (Shanghai, China). Bovine serum albumin (BSA) buffer (5%) was bought from Solarbio (Beijing, China). Rabbit anti-IL-10 polyclonal antibody (bs-20373R), rabbit anti-IL-4 polyclonal antibody (bs-0581R) and rabbit anti-IFN-γ polyclonal antibody (bs-0480R) were obtained from Bioss antibodies (Beijing, China). Rabbit anti-TNF-α polyclonal antibody (BA14901), HRP-conjugated polymer anti-rabbit IgG (SV0002) and hematoxylin solution (AR1180-1) were bought from Boster Biological Technology (Wuhan, Hubei, China). SuperSignal® west pico chemiluminescent substrate was purchased from ThermoFisher Scientific (Waltham, MA). TNBS was bought from Sigma (St. Louis, MO). Ultrapure RNA kit (CW0581M), HiFiScript cDNA synthesis kit (CW2569M), UltraSYBR mixture (CW0957M), BCA kit (CW0014S), and diaminobenzidine (DAB) kit (CW0125) were obtained from CWBio (Beijing, China).
Sixteen male C57 mice (weight 20–24 g, 8 weeks old) were purchased from Changzhou Cavens Laboratory Animal Co., Ltd. (license no.: SCXK(Su)2016–0010; Jiangsu, China). Mice were raised in an environment with 18–29 °C, 40–70% relative humidity, and 12h/12h light/dark cycle. They had free access to food and water. Animal protocols were approved by the Ethics Committee of the First Affiliated Hospital of Wenzhou Medical University.
IBD mice model establishment
IBD mice model was established according to a previous report [Citation11]. In brief, mice fasted for 36 h and were mildly anesthetized with ethyl ether. Mice were then kept in the prone position. The cannula of 18 G venous indwelling needle was gently inserted into the colon through the anus of mice about 4 cm from the tip of the cannula to the anus. A trocar connecting a 1 mL syringe was inserted immediately into the cannula and the mice were made upside down. 100 µL 50% ethanol containing 1.5 mg TNBS was slowly injected. The trocar was pulled out. The mice were kept upside down for 30 s and put back into the cage.
Animal grouping and sampling
The mice were divided into four groups (n = 4): the control group, the model group, Ad5 group, and Ad5-CDC42 group. Mice in the control group did not suffer the IBD modeling operation. Mice in the model group suffered the IBD modeling operation. After the IBD modeling operation, mice which were treated with adenovirus empty vector and CDC42 adenovirus expression vector served as the Ad5 group and Ad5-CDC42 group, respectively.
Two days after IBD modeling operation, 20 µL adenovirus empty vector and CDC42 adenovirus expression vector were intravenously injected into the mice in the Ad5 group and the Ad5-CDC42 group, respectively [Citation12]. Mice in the model group were administrated with 20 µL normal saline. After 3 days, the mice were anesthetized and the chest was opened to collect blood from the heart for ELISA assay. The abdominal cavity was opened afterwards and the descending colon was collected for RT-PCR, western blot, and immunohistochemical analysis.
RT-PCR
Total RNA was extracted from the colon samples by using the Ultrapure RNA kit according to the manufacturer’s instruction. RNA concentration and purity were determined by using Nanodrop 2000 (Thermo Fisher Scientific, Waltham, MA). RNA was reversely transcribed into cDNA by using the HiFiScript cDNA synthesis kit according to the manufacturer’s instruction. Primers were prepared by General Biosystems Inc. (Anhui, China) and provided in . The PCR reaction system was composed of 10 µL 2× UltraSYBR Mixture, 0.8 µL forward/reverse Primers (10 mM), 0.4 µL 50× ROX reference dye II, and 2 µL cDNA template. The reaction parameters were as follows: initial denaturation at 95 °C for 30 s, denaturation at 95 °C for 3 s, annealing at the temperature shown in for 30 s, elongation for 30 s at 72 °C for 40 circles. The amplification products were separated through agarose gel electrophoresis. The dissociation curve was analyzed as follows: 15 s at 95 °C, 1 min at the temperature shown in , 15 s at 95 °C, 15 s at the temperature shown in , 15 s at the temperature shown in , and measured stepwise from 95 °C every 0.5 °C. Eventually, it was evaluated on a fluorescent quantitation PCR (CFX Connect™, Bio-Rad). GAPDH served as internal control. Relative expression levels of genes were calculated by using the 2–ΔΔCt method [Citation13].
Table 1. The primers of RT-PCR.
Western blot
The colon samples were ground and lysed in lysis buffer in an ice bath for 30 min. The lysate was centrifuged at 10,000 rpm and 4 °C for 10 min. The supernatant was carefully collected to obtain total protein. Protein concentration was quantified by using BCA kit. Proteins were separated by SDS-PAGE for 2 h and transferred to polyvinylidene difluoride membrane by a wet method for 50 min. After incubation in 5% BSA buffer at room temperature for 1 h, the membrane was incubated in primary antibody buffer at 4 °C overnight. The membrane was then rinsed three times with Tris buffered saline with Tween 20 (TBST) and incubated in secondary antibody buffer at room temperature for 2 h. SuperSignal® west pico chemiluminescent substrate was dropwise added onto the membrane which was thereafter examined on a gel imaging system (ChemiDoc™ XRS+, Bio-Rad). Gray values were analyzed by using Quantity One software (v4.62, Bio-Rad). GAPDH served as internal control as well.
Immunohistochemical analysis
The colon samples were embedded with paraffin and cut into slices. The slices were baked, dewaxed, hydrated, incubated in citric acid buffer and boiled under high pressure to repair the antigen. The slices were then transferred to a wet box, and fresh 3% hydrogen peroxide was added to remove endogenous peroxidase. 5% BSA buffer was dropwise added onto the slices for non-specific blocking. Afterwards, the slices were incubated in primary antibody buffer at 4 °C overnight. After washing, they were incubated in secondary antibody buffer at 37 °C for 1 h. After washing again, the slices were developed with DAB kit for 5–10 min. After washing again, they were stained in hematoxylin solution for 3 min. Finally, the slices were differentiated in acidic alcohol, blued in bluing buffer, washed, dehydrated, transparentized, mounted and examined by a microscope (CX41, Olympus, Japan).
ELISA assay
The blood was centrifuged at 3000 rpm for 5 min to obtain the supernatant. The contents of IL-4, IL-10, TNF-α, and IFN-γ in the supernatant were determined by their ELISA kits according to their manufacturer’s instructions. Absorbance was measured within 15 min after adding the stop solution at 450 nm on a microplate reader (RT-6100, Rayto). The contents were calculated through the standard curve.
Statistical analysis
Values of mean and standard deviation were used to summarize continuous variables. Statistical analysis was performed by using SPSS software (v17.0, SPSS Inc.). One-way analysis of variance (AVONA) and least significant difference post hoc-test were used to assess the differences. p < .05 was considered to indicate a statistically significant difference.
Results
mRNA levels of IL-4, IL-10, TNF-α, and IFN-γ in colon tissues
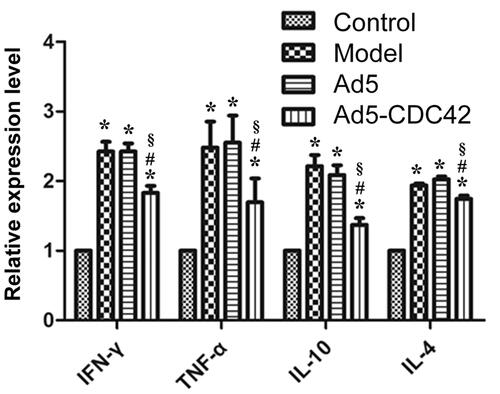
The mRNA levels of IL-4, IL-10, TNF-α, and IFN-γ in the colon tissues of various groups which were evaluated by RT-PCR were shown in . Their mRNA levels were lowest in the control group and their mRNA levels in the model group, the Ad5 group and the Ad5-CDC42 group were remarkably higher than those in the control group (p < .05). Their mRNA levels in the model group and the Ad5 group were similar (p > .05). Compared with the model group and the Ad5 group, their mRNA levels were significantly reduced in the Ad5-CDC42 group (p < .05).
Figure 1. The mRNA levels of IL-4, IL-10, TNF-α, and IFN-γ in the colon tissues of various groups which were evaluated by RT-PCR. Control: the control group; Model: the model group; Ad5: the Ad5 group (adenovirus empty vector); Ad5-CDC42: the Ad5-CDC42 group (CDC42 adenovirus expression vector). *p < .05 vs. control. #p < .05 vs. model. §p < .05 vs. Ad5.
Protein levels of IL-4, IL-10, TNF-α, and IFN-γ in colon tissues
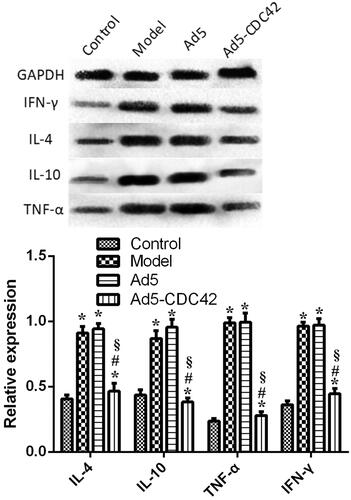
The protein levels of IL-4, IL-10, TNF-α, and IFN-γ in the colon tissues of various groups which were assessed by western blot were displayed in . Interestingly, the protein levels of IL-4, IL-10, TNF-α, and IFN-γ were comparable between the control group and the Ad5-CDC42 group, and their protein levels were similar between the model group and the Ad5 group (p > .05). Compared with the model group and the Ad5 group, their protein levels were notably lowered in the Ad5-CDC42 group (p < .05).
Figure 2. The protein levels of IL-4, IL-10, TNF-α, and IFN-γ in the colon tissues of various groups which were assessed by western blot. Control: the control group; Model: the model group; Ad5: the Ad5 group (adenovirus empty vector); Ad5-CDC42: the Ad5-CDC42 group (CDC42 adenovirus expression vector). *p < .05 vs. control. #p < .05 vs. model. §p < .05 vs. Ad5.
Contents of IL-4, IL-10, TNF-α, and IFN-γ in serum
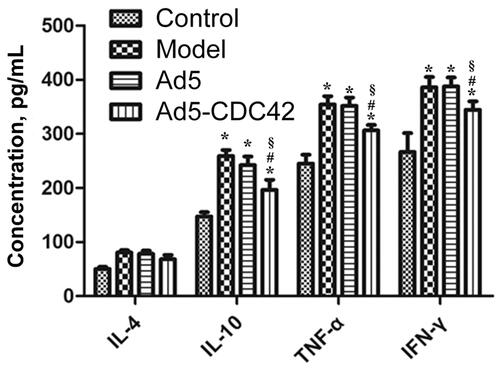
The contents of IL-4, IL-10, TNF-α, and IFN-γ in the serum of various groups which were determined by ELISA assay were shown in . Their contents were lowest in the control group and their contents in the model group, the Ad5 group and the Ad5-CDC42 group were significantly higher than those in the control group (p < .05). Their contents in the model group and the Ad5 group were similar (p > .05). In comparison with the model group and the Ad5 group, their contents were significantly decreased in the Ad5-CDC42 group (p < .05).
Figure 3. The contents of IL-4, IL-10, TNF-α, and IFN-γ in the serum of various groups which were determined by ELISA assay. Control: the control group; Model: the model group; Ad5: the Ad5 group (adenovirus empty vector); Ad5-CDC42: the Ad5-CDC42 group (CDC42 adenovirus expression vector). *p < .05 vs. control. #p < .05 vs. Model. §p < .05 vs. Ad5.
Immunohistochemical analysis of IL-4, IL-10, TNF-α, and IFN-γ in colon tissues
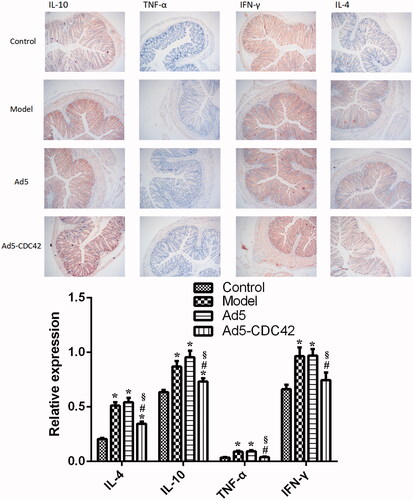
depicted the immunohistochemical results of IL-4, IL-10, TNF-α, and IFN-γ in the colon tissues of various groups. Their levels were lowest in the control group. Their levels in the model group and the Ad5 group were similar (p > .05) and significantly higher than those in the control group (p < .05). In comparison with the model group and the Ad5 group, their levels were significantly reduced in the Ad5-CDC42 group (p < .05).
Figure 4. The immunohistochemical results of IL-4, IL-10, TNF-α, and IFN-γ in the colon tissues of various groups. Target proteins were displayed in brown. Magnification × 100. Control: the control group; Model: the model group; Ad5: the Ad5 group (adenovirus empty vector); Ad5-CDC42: the Ad5-CDC42 group (CDC42 adenovirus expression vector). *p < .05 vs. control. #p < .05 vs. Model. §p < .05 vs. Ad5.
Discussion
Intestinal epithelial damage is the most direct pathogenic factor of IBD [Citation1]. Intestinal epithelial damage causes a large number of antigenic substances to enter the body, triggering innate immune response; When the innate immune response system is dysfunctional, acute or chronic inflammatory response regulated by signaling pathways such as Toll-like receptor (TLR) will be abnormal, in which will induce IBD such as colitis [Citation2]. IL-10, IL-4, TNF-α, and IFN-γ are interrelated cytokines involved in the regulation of a large number of innate immune responses [Citation14]. IL-10 down-regulates the expression of Th1 cytokines and major histocompatibility complex (MHC) II antigen, participates in the regulation of macrophage’s development and transformation, and promotes the proliferation and growth of B cells [Citation15,Citation16]. IL-4 can induce Th0 cells to differentiate into Th2 cells, while Th2 cells can produce IL-4, forming a positive feedback; In addition, IL-4 enhances the M2 stimulation of macrophages through the downstream signal transducers and activators of transcription 6 (STAT6) [Citation17,Citation18]. TNF-α is produced primarily by activated macrophages, and its endogenous pyrogen; TNF-α can induce acute phase reactions and the TNF-α imbalance is closely associated with IBD [Citation19,Citation20]. IFN-γ has the ability to directly inhibit virus replication; More importantly, IFN-γ is an important activator of macrophages and can up-regulate the expression of MHC II antigen [Citation21]. In this study, the functions of these four cytokines IL-10, IL-4, TNF-α, and IFN-γ were mutually regulated.
CDC42 overexpression enhances the migration of macrophages to damaged intestinal mucosa and increases the activity of macrophages, thus promoting the treatment of IBD. In this study, the expression of CDC42 in IBD mice was increased in a short time by injecting adenovirus packaged with CDC42 expression vector into the tail vein of IBD mice. After 3 days, we performed diverse tests such as RT-PCR, western blot, immunohistochemical analysis, and ELISA assay and revealed that the levels of IL-10, IL-4, TNF-α, and IFN-γ were elevated in the colon tissues and serum of IBD mice, while they were remarkably reduced by CDC42 treatment. It was suggested that CDC42 could regulate the inflammatory response in IBD mice and the increased expression of CDC42 could accelerate the development and rehabilitation of the innate immune response and inflammatory response.
One of the key factors to produce this effect may be the regulation of CDC42 on macrophages. Macrophages are the crucial cells in innate immune response. Their development, transformation, and function are regulated by many signal proteins, and they are also the producer of many cytokines [Citation22–24]. In addition, macrophages have a strong ability of producing pseudopodia, phagocytosis, and producing vesicles, all of which are associated with CDC42 [Citation22–24]. Besides, the down-regulation of IL-10, IL-4, TNF-α, and IFN-γ expression in IBD mice induced by CDC42 administration is possibly resulted from the sampling time which is assigned at 3 days after CDC42 administration. At this time point, the innate immune response has reached the stage of rehabilitation. In order to understand the changes of cytokines expression and the effect of CDC42 on innate immune response more accurately and particularly, in the future, we will improve the experimental design, arrange multiple time intervals and directly examine the macrophages.
In conclusion, the levels of inflammatory cytokines IL-10, IL-4, TNF-α, and IFN-γ were elevated in the colon tissues and serum of IBD mice, which could be reduced by the CDC42 treatment through intravenous injection of CDC42 adenovirus expression vector. CDC42 regulated the inflammatory response and the innate immune response in IBD mice. These results may provide some promising insights into drug development and clinical diagnosis for IBD therapy.
Disclosure statement
The authors report no conflict of interest.
Data availability
The analyzed data sets generated during the study are available from the corresponding author on reasonable request.
Additional information
Funding
References
- Maloy KJ, Powrie F. Intestinal homeostasis and its breakdown in inflammatory bowel disease. Nature. 2011;474:298–306.
- Cario E. Toll-like receptors in inflammatory bowel diseases: a decade later. Inflamm Bowel Dis. 2010;16:1583–1597.
- Mills CD. M1 and M2 macrophages: oracles of health and disease. Crit Rev Immunol. 2012;32:463–488.
- Stengel K, Zheng Y. Cdc42 in oncogenic transformation, invasion, and tumorigenesis. Cell Signal. 2011;23:1415–1423.
- Shiraishi A, Uruno T, Sanematsu F, et al. DOCK8 protein regulates macrophage migration through Cdc42 protein activation and LRAP35a protein interaction. J Biol Chem. 2017;292:2191–2202.
- Mohammadi S, Isberg RR. Cdc42 interacts with the exocyst complex to promote phagocytosis. J Cell Biol. 2013;200:81–93.
- Cerione RA. Cdc42: new roads to travel. Trends Cell Biol. 2004;14:127–132.
- Sinha S, Yang W. Cellular signaling for activation of Rho GTPase Cdc42. Cell Signal. 2008;20:1927–1934.
- Erickson JW, Cerione RA. Multiple roles for Cdc42 in cell regulation. Curr Opin Cell Biol. 2001;13:153–157.
- Pizarro TT, Arseneau KO, Bamias G, et al. Mouse models for the study of Crohn's disease. Trends Mol Med. 2003;9:218–222.
- te Velde AA, Verstege MI, Hommes DW. Critical appraisal of the current practice in murine TNBS-induced colitis. Inflamm Bowel Dis. 2006;12:995–999.
- Liu Q, Muruve DA. Molecular basis of the inflammatory response to adenovirus vectors. Gene Ther. 2003;10:935–940.
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001;25:402–408.
- Martinez FO, Gordon S. The M1 and M2 paradigm of macrophage activation: time for reassessment. F1000Prime Rep. 2014;6:13.
- Moore KW, De Waal Malefyt R, Coffman RL, et al. Interleukin-10 and the interleukin-10 receptor. Annu Rev Immunol. 2001;19:683–765.
- Akdis CA, Joss A, Akdis M, et al. A molecular basis for T cell suppression by IL-10: CD28-associated IL-10 receptor inhibits CD28 tyrosine phosphorylation and phosphatidylinositol 3-kinase binding. FASEB J. 2000;14:1666–1668.
- Van Dyken SJ, Locksley RM. Interleukin-4 and interleukin-13-mediated alternatively activated macrophages: roles in homeostasis and disease. Annu Rev Immunol. 2013;31:317–343.
- Sokol CL, Barton GM, Farr AG, et al. A mechanism for the initiation of allergen-induced T helper type 2 responses. Nat Immunol. 2008;9:310–318.
- Schoenborn JR, Wilson CB. Regulation of interferon-gamma during innate and adaptive immune responses. Adv Immunol. 2007;96:41–101.
- Chesler DA, Reiss CS. The role of IFN-gamma in immune responses to viral infections of the central nervous system. Cytokine Growth Factor Rev. 2002;13:441–454.
- Zhao H, Liao X, Kang Y. Tregs: where we are and what comes next? Front Immunol. 2017;8:1578.
- Wynn TA, Chawla A, Pollard JW. Macrophage biology in development, homeostasis and disease. Nature 2013;496:445–455.
- Sica A, Mantovani A. Macrophage plasticity and polarization: in vivo veritas. J Clin Invest. 2012;122:787–795.
- Okabe Y, Medzhitov R. Tissue biology perspective on macrophages. Nat Immunol. 2016;17:9–17.
