Abstract
LncRNA SBF2-AS1 has been reported to be implicated in the deterioration of multiple human cancers. However, the roles and underlying mechanisms of SBF2-AS1 in acute myeloid leukemia (AML) are still unclear. In the present study, the online GEPIA database showed that SBF2-AS1 expression was significantly increased in AML samples. QRT-PCR results showed that SBF2-AS1 expression was upregulated in AML cells. CCK-8 assay revealed that SBF2-AS1 inhibition decreased AML cells proliferation ability in vitro. Flow cytometry assays showed that SBF2-AS1 inhibition induced AML cells apoptosis and arrested AML cells in G0/G1 phase. Mechanistically, miR-188-5p was identified as a direct target of SBF2-AS1. SBF2-AS1 upregulated the expression level of ZFP91 by sponging miR-188-5p. And the effects of SBF2-AS1 suppression on AML cells progression could be abolished by miR-188-5p inhibitors. Moreover, we found that SBF2-AS1 inhibition reduced tumor growth in vivo. Taken together, our findings elucidated that SBF2-AS1 could act as a miRNA sponge in AML progression, and provided a potential therapeutic strategy for AML treatment.
Introduction
Acute myeloid leukemia (AML) is recognized as one of the most common hematologic malignancies worldwide [Citation1]. Although various strategies (new chemotherapeutic agents, etc.) have been applied to control AML, 1/3 to 1/2 of children with AML continue to relapse [Citation2,Citation3]. Thus, a more complete understanding of the molecular mechanisms becomes essential for development effective therapeutic targets against AML.
Long non-coding RNAs (lncRNAs) are a non-coding RNA of more than 200 nt in length and devoid of coding protein functions [Citation4]. However, recent studies showed that lncRNAs might exert important regulating effects on epigenetic modification, transcription and post-transcriptional processing [Citation5,Citation6]. Recently, studies showed that aberrant expression of lncRNAs could play vital roles in tumorigenesis and development of various cancers [Citation7], including AML. For example, Ma et al. revealed that lncRNA LINC00265 predicted the prognosis of acute myeloid leukemia patients and functioned as a promoter by activating PI3K-AKT pathway [Citation8]. Sun et al. found that lncRNA ANRIL regulated AML development through modulating the glucose metabolism pathway of AdipoR1/AMPK/SIRT1 [Citation9]. Dong et al. revealed that lncRNA HOXA-AS2 inhibition suppressed chemoresistance of AML via the miR-520c-3p/S100A4 Axis [Citation10].
SBF2-AS1 is a 2708 nt antisense RNA to SBF2, which is located at the 11p15.1 locus [Citation11]. Recent studies showed that SBF2-AS1 might play critical roles in tumor progression, such as lung cancer, cervical cancer, esophageal squamous cell carcinoma, and liver cancer [Citation12–15]. However, the roles and underlying mechanisms of SBF2-AS1 in AML remain unclear. In the present study, we revealed that SBF2-AS1 could act as an oncogenic lncRNA in AML progression. Mechanistically, SBF2-AS1 could sponge miR-188-5p as a ceRNA to upregulate ZFP91 expression, thus accelerating AML cells’ proliferation. Thus, our data suggested that SBF2-AS1 could act is an oncogenic lncRNA and facilitated the proliferation of AML mainly by regulating the miR‐188‐5p/ZFP91 signaling axis.
Materials and methods
Cell culture and transfection
Human bone marrow stromal cell line HS-5 and AML cell lines (THP‐1, U937, HL60) were purchased from ATCC (Manassas, VA). The cells were cultured in a RPMI1640 medium containing 10% fetal bovine serum (FBS) and 100 U/mL penicillin and 100 μg/mL streptomycin. Cells were maintained with 5% CO2 at 37 °C.
Small interfering RNAs (siRNAs) specific targeting SBF2-AS1 (si-SBF2-AS1), miR‐188‐5p mimic, miR‐188‐5p inhibitor, or negative control were all purchased from GenePharma (Shanghai, China). Cell transfection was by using Lipofectamine 2000 according (Invitrogen) to the manufacturer’s protocols. The siRNA sequences for SBF2-AS1 were as following: si-SBF2-AS1-1 5′-CAGAAGGAGUCUACUGCUAAG-3′ (Sense) and 5′-UAGCAGUAGACUCCUUCUGGG-3′ (Antisense); si-SBF2-AS1-2 5′-GCAAGCCUGCAUGGUACAUTT -3′ (Sense) and 5′-AUGUACCAUGCAGGCUUGCTT-3′ [Citation12].
Cell proliferation assay
Cell proliferation was assessed using the Cell Counting Kit-8 according to the manufacturer's protocols (CCK-8, Dojindo, Japan). Briefly, the logarithmically growing cells (1 × 104/well) were inoculated into 96-well culture plates and cultured for indicated times. Then 10 μL CCK-8 solutions were added and incubated for 2 h at 37 °C. Absorbance at 450 nm was measured using a microtiter plate reader (Bio-Rad, CA) [Citation13].
Flow cytometry assay
The apoptosis of cells was explored by Annexin V-FITC/PI Apoptosis Detection Kit (Sigma, Louis, MO). At 48 h post-transfection, cells were resuspended in Annexin-binding buffer and double stained with Annexin V/FITC (5 μL) and PI solution (10 μL) at 37 °C for 15 min in darkness. Following the addition of PBS, cells were analyzed by flow cytometry (BD Biosciences).
For cell cycle detection, transfected cells were treated with 0.25% trypsin, fixed in 70% chilled ethanol at 4 °C overnight, and centrifuged at 1000 r/min. Cell suspension was then added with 10 μL RNase and stained with 1% PI in the dark for 30 min. The fow cytometry (FACSCalibur, BD Biosciences) was used to detect cell cycle.
RNA extraction and quantitative real-time PCR (qRT-PCR)
Total RNA was extracted from tissues and cultured cells using TRIZOL reagents according to the manufacturer's protocols (Invitrogen, Carlsbad, CA). For qRT-PCR, the isolated RNA was reverse transcribed to cDNA using a Reverse Transcription Kit (Takara, China). Real-time PCR analyses were conducted with StepOne Plus Real-Time PCR system (Applied Biosystems, CA) with SYBR® Premix Ex Taq™ Reagent (TaKaRa, Japan). QRT-PCR was performed at 95 °C for 5 min, 95 °C for 15 s, 58 °C for 30 s, and 74 °C for 30 s, for a total of 40 cycles [Citation16]. The primers used in this study were as following: SBF2-AS1 (forward): 5′-CACGACCCAGAAGGAGTCTAC-3′, and (reverse): 5′-CCCGGTACCTTCCTGTCATA-3′.
Western blot
The method used in the study is according to the previous study [Citation16].
Luciferase reporter assay
The wild-type (WT) sequence of SBF2-AS1 and the WT sequence of ZFP91 3’UTR containing the putative binding region for miR‐188‐5p were amplified and fused into the pGL3-control vector (Promega, Madison, WI) to generate the SBF2-AS1-WT and ZFP91-WT constructs, respectively. Correspondingly, the mutant (MUT) sequences of SBF2-AS1 and ZFP91 were introduced to create the SBF2-AS1-MUT and ZFP91-MUT constructs, respectively. These constructions were transfected into cells, together with miR‐188‐5p mimics and miR-NC. The relative luciferase activity was measured 48 h post-transfection using the dual-luciferase reporter assay (Promega) by following the manufacturer’s instructions.
RNA immunoprecipitation (RIP) assay
RNA immunoprecipitation experiments were performed by using Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore) with AGO2 antibody following the manufacturer’s instructions and previous study [Citation17].
In vivo xenograft assay
The female BALB/c-nu mice (3–4 weeks old) were obtained from the Chinese Academy of Medical Sciences (Beijing, China) and manipulated according to protocols approved by the Animal Care Committee of The First Affiliated Hospital of Xinxiang Medical University. Transfected cells were subcutaneously injected into the left axilla of nude mice. Tumor volume was measured by a Vernier caliper every week. Tumor volume was calculated as (A × B2)/2, where A was the long diameter and B was the short diameter of the transplanted tumor.
Statistical analysis
SPSS 19.0 was used for all statistical analysis. Data were represented as mean ± SD. Statistical significance was analyzed by student’s t-test or one-way analysis of variance. p < .05 indicated the significant difference.
Results
SBF2-AS1 was up-regulated in AML
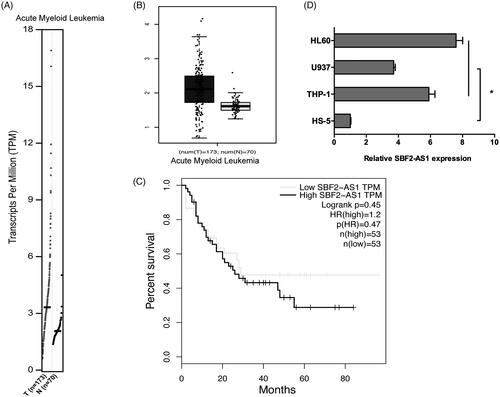
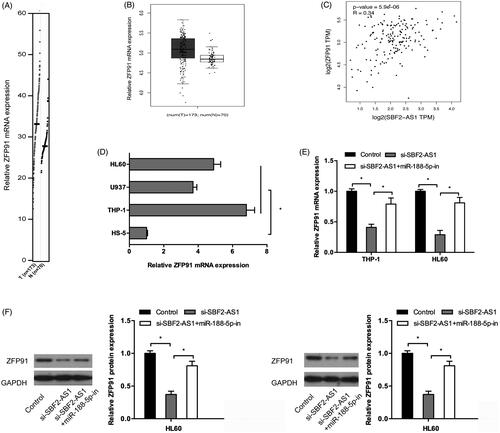
To explore the roles of SBF2-AS1 in AML progression, we firstly determined SBF2-AS1 expression in AML by GEPIA database. Results showed that SBF2-AS1 mRNA expression was significantly increased in AML samples (n = 173) compared to normal samples (n = 70) (). GEPIA data revealed that high SBF2-AS1 expression was associated with poor overall survival in AML patients (). In addition, we determined SBF2-AS1 expression in AML cells, qRT-PCR results showed that SBF2-AS1 expression was significantly increased in AML cell lines (THP‐1, U937, and HL60) compared to HS-5 cells (). Those data indicated that SBF2-AS1 might play critical roles in AML progression.
Figure 1. High expression of SBF2-AS1 in AML. (A, B) SBF2-AS1 expression in AML samples (n = 173) and normal samples (n = 70) from GEPIA analysis. (C) The survival curves of AML patients from GEPIA analysis. (D) qRT-PCR analysis for SBF2-AS1 expression in AML cell lines (THP‐1, U937 and HL60). *p < .05.
SBF2-AS1 inhibition reduced AML cells proliferation
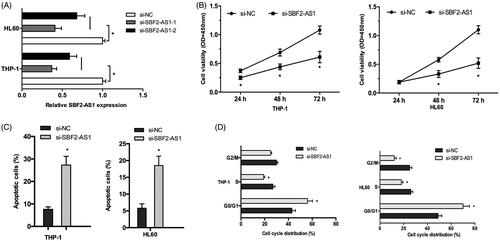
To identify functions of SBF2-AS1 in AML cells, we knocked-down SBF2-AS1 expression in THP‐1 and HL60 cells and choose si-SBF2-AS1-1 for the subsequent experiments (). CCK-8 assay revealed that SBF2-AS1 suppression significantly reduced the proliferation ability of AML cells (). Flow cytometry assays were used to explore the effects of SBF2-AS1 on AML cells apoptosis and cell cycle. Cells apoptosis assay showed that SBF2-AS1 inhibition noticeably increased the apoptosis rate in THP‐1 and HL60 cells (). Cell cycle assay revealed that SBF2-AS1 suppression significantly arrested THP‐1 and HL60 cells in G0/G1 phase (). Those data indicated that SBF2-AS1 inhibition reduced AML cells proliferation ability.
Figure 2. SBF2-AS1 inhibition decreases AML cells proliferation. (A) qRT-PCR analysis for SBF2-AS1 knockout efficiency in THP‐1 and HL60 cells. (B) CCK-8 assay showed that si-SBF2-AS1 significantly decrease AML cells proliferation ability. (C) Flow cytometry analysis revealed that SBF2-AS1 inhibition induced AML cells apoptosis. (D) Flow cytometry analysis showed that SBF2-AS1 inhibition arrested AML cells in G0/G1 phase. *p < .05.
MiR‐188‐5p is a target of SBF2-AS1
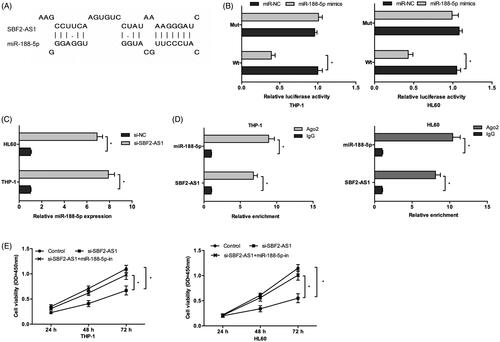
Increasing studies indicated that lncRNAs could bind to miRNAs and regulate the activity and expression of miRNAs [Citation18]. In the present study, we revealed that that SBF2-AS1 might interact with miR-188-5p (). Dual-luciferase assay showed that the luciferase activity was significantly decreased by co-transfection with miR-188-5p mimics and SBF2-AS1-Wt vector (). QRT-PCR showed that SBF2-AS1 inhibition significantly increased miR-188-5p expression in AML cells (). RIP assay revealed that both SBF2-AS1 and miR-188-5p expression was highly enriched in Ago2 immunoprecipitates relative to control IgG immunoprecipitates (). Moreover, CCK-8 assay found that miR-188-5p inhibitors significantly reversed the effects of SBF2-AS1 knockdown on AML cells proliferation (). Thus, these data indicated that SBF2-AS1 might influence AML cells physiology in a miR-188-5p-dependent manner.
Figure 3. SBF2-AS1 binds to miR-188-5p in AML. (A) The schematic diagram of SBF2-AS1 with miR-188-5p binding sites. (B) Luciferase reporter assay showed that luciferase activity was significantly decreased by co-transfection with miR-188-5p mimics and SBF2-AS1-Wt vector. (C) QRT-PCR showed that SBF2-AS1 inhibition decreased miR-188-5p expression in AML cells. (D) RIP assay revealed that both SBF2-AS1 and miR-188-5p expression were highly enriched in Ago2 immunoprecipitates. (E) CCK-8 assay showed that mIR-188-5p inhibitors reversed the effects of SBF2-AS1 knockdown on AML cells proliferation. *p < .05.
ZFP91 is a direct target of miR-188-5p
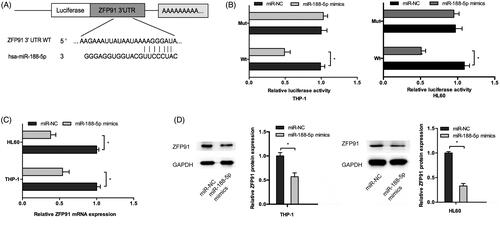
Next, we determined the mechanisms underlying the activity of miR-188-5p in AML, TargetScan and miRBase showed that ZFP91 was predicted as a putative target of miR-188-5p (). Dual-luciferase assay showed that miR-188-5p mimics significantly decreased the luciferase activity of ZFP91-Wt but not ZFP91-Mut (). QRT-PCR showed that miR-188-5p mimics significantly reduced ZFP91 mRNA expression in AML cells (). Western blot showed a similar result as qRT-PCR (). Thus, these results indicated that ZFP91 is a direct target gene of miR-188-5p in AML cells.
Figure 4. ZFP91 is a direct target of miR-188-5p. (A) The schematic diagram of ZFP91 with miR-188-5p binding sites. (B) Luciferase reporter assay showed that miR-188-5p mimics significantly decreased the luciferase activity of ZFP91-Wt but not ZFP91-Mut. (C, D) MiR-188-5p mimics greatly downregulated ZFP91 expression in AML cells. *p < .05.
SBF2-AS1 modulates ZFP91 activity through miR-188-5p
We further sought to the relationship between ZFP91 and SBF2-AS1 in AML. GEPIA database showed that ZFP91 expression was significantly upregulated and correlated with SBF2-AS1 expression in AML samples (). QRT-PCR showed that ZFP91 expression was significantly increased in AML cell lines (THP‐1, U937 and HL60) compared to HS-5 cells (). Moreover, we found that SBF2-AS1 inhibition significantly decreased ZFP91 expression both at mRNA and protein levels in AML cells, and the inhibition effects could be abolished by miR-188-5p inhibitors (). These data indicated that SBF2-AS1 could activity ZFP91 by regulating miR-188-5p expression.
Figure 5. SBF2-AS1 modulates ZFP91 activity through miR-188-5p. (A, B) ZFP91 expression in AML samples (n = 173) and normal samples (n = 70) from GEPIA analysis. (C) ZFP91 expression was positively correlated with SBF2-AS1 expression in AML samples. (D) QRT-PCR analysis for ZFP91 expression in AML cell lines (THP‐1, U937, and HL60). (E, F) SBF2-AS1 inhibition significantly decreased ZFP91 expression both in mRNA and protein levels in AML cells, and the inhibitory effects could be reversed by miR-188-5p inhibitors. *p < .05.
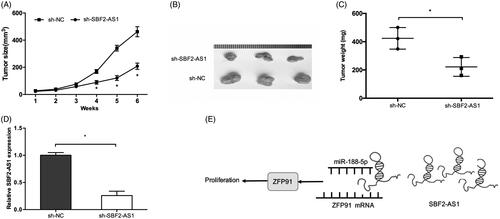
SBF2-AS1 promoted tumor growth in vivo
We further studied the effects of SBF2-AS1 in xenograft tumor mice in vivo. Results showed that tumors formed in the sh-SBF2-AS1 group were dramatically smaller than that in sh-NC group (). Consistently, tumor growth and average tumor weight in sh-SBF2-AS1 group was significantly lower than that in sh-NC group (). Furthermore, SBF2-AS1 expression was determined in xenograft tumor tissues. We found that SBF2-AS1 expression was significantly inhibited in sh-SBF2-AS1 group (). To sum up, these suggested that increased SBF2-AS1 in AML reduces the activity of miR-188-5p to increase ZFP91 expression, and finally promoting ALM proliferation ().
Figure 6. SBF2-AS1 promotes tumor growth in vivo. (A, B) SBF2-AS1 silencing suppressed tumor growth in vivo. (C) The tumor weight of nude mice was measured. (D) The expression level of SBF2-AS1 in tumors of nude mice was detected by qRT-PCR. (E) A schematic diagram of SBF2-AS1/miR-188-5p/ZFP91 regulatory pathway in AML. *p < .05.
Discussion
Accumulating evidence demonstrated that lncRNAs were involved in the inactivation of tumor suppressor genes or activation of oncogenes mediated tumor progression. For example, Zhang et al. showed that upregulation of lncRNA MALAT1 correlated with tumor progression and poor prognosis in clear cell renal cell carcinoma [Citation19]. Li et al. revealed that lncRNA CASC2 suppressed the proliferation of gastric cancer cells by regulating the MAPK signaling pathway [Citation20]. Chen et al. found that lncRNA CCAT1 promoted multiple myeloma progression by acting as a molecular sponge of miR-181a-5p to modulate HOXA1 expression [Citation21]. In the present study, we found that SBF2-AS expression was highly expressed in AML samples. SBF2-AS suppression significantly decreased AML cells proliferation in vitro and reduced tumor growth in vivo. Mechanistically, SBF2-AS reduced miR-188-5p-mediated repression of ZFP91, thereby facilitating the proliferation of AML cells.
Recent studies showed that lncRNAs might serve as a ceRNA to compete for miRNAs, thereby negatively regulating the expression of miRNAs [Citation18]. For example, Xu et al. revealed that lncRNA DANCR functioned as a ceRNA to regulate RAB1A expression by sponging miR-634 in glioma [Citation22]. Gao et al. found that lncRNA FLVCR1-AS1 contributed to the proliferation and invasion of lung cancer by sponging miR-573 to upregulate E2F3 expression [Citation23]. In the present study, we also found that SBF2-AS could function as microRNA sponge. The results of luciferase reporter and RNA pull-down assays showed that miR-188-5p was a direct target of SBF2-AS. MiR-188-5p has been identified to be significantly decreased in multiple cancers, including hepatocellular carcinoma [Citation24], gastric cancer [Citation25], and prostate cancer [Citation26]. Likewise, our data also confirmed that miR-188-5p was a tumor suppressor in AML. Thus, SBF2-AS might exert the growth-promoting role mainly by binding and inhibiting the tumor suppressor miR-188-5p.
It has been well documented that miRNA could master the expression of the gene by post-transcriptional levels [Citation27]. Here, we found that miR-188-5p could directly interact with the 3′-UTR of Zinc finger protein 91 (ZFP91) mRNA to suppress ZFP91 expression. Zinc finger protein 91 (ZFP91), a conserved 63.5 kDa nuclear protein with structural motifs characteristic of transcription factors [Citation28]. Increasing evidence showed that ZFP91 could play critical roles in tumor progression. For example, Tompkins et al. found that ZFP91 interacted with the ARF tumor suppressor, which is known for its induction of p53-dependent cell death or growth arrest in response to activated oncogenes [Citation29]. Jin et al. showed that ZFP91 might play a key role in LIGHT-induced activation of non-canonical NF-κB pathway [Citation30]. Ma et al. found that ZFP91 activated HIF-1α via NF-κB/p65 to promote proliferation and tumorigenesis of colon cancer [Citation31].
Recently, Unoki et al. found that ZFP91 was significantly increased in AML, ZFP91 inhibition could significantly suppress AML cells growth, and induce apoptosis [Citation32]. In the present study, we showed that ZFP91 was significantly increased in AML samples and cell lines. High ZFP91 expression was associated with poor overall survival of AML patients. Moreover, we found that SBF2-AS1 inhibition significantly decreased ZFP91 expression both at mRNA and protein levels in AML cells, and the inhibition effects could be abolished by miR-188-5p inhibitors. Thus, these data indicated that the regulatory network of SBF2-AS1/miR-188-5p/ZFP91 could play an essential role in AML progression.
In conclusion, we suggested that SBF2-AS1 was an oncogenic lncRNA and facilitated the growth of AML mainly by regulating the miR-188-5p/ZFP91 signaling axis, which provides a potential therapeutic target for AML patients treatment.
Disclosure statement
The authors declare that they have no competing interests.
References
- Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. Cancer J Clin. 2015;65:87–108.
- Deschler B, Lübbert M. Acute myeloid leukemia: epidemiology and etiology. Cancer 2006;107:2099–2107.
- Pui CH, Ribeiro RC, Hancock ML, et al. Acute myeloid leukemia in children treated with epipodophyllotoxins for acute lymphoblastic leukemia. N Engl J Med. 1991;325:1682–1687.
- Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell 2009;136:629–641.
- Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell 2011;43:904–914.
- Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–361.
- Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell 2016;29:452–463.
- Ma L, Kuai WX, Sun XZ, et al. Long noncoding RNA LINC00265 predicts the prognosis of acute myeloid leukemia patients and functions as a promoter by activating PI3K-AKT pathway. Eur Rev Med Pharmacol Sci. 2018;22:7867–7876.
- Sun LY, Li XJ, Sun YM, et al. LncRNA ANRIL regulates AML development through modulating the glucose metabolism pathway of AdipoR1/AMPK/SIRT1. Mol Cancer 2018;17:127.
- Dong X, Fang Z, Yu M, et al. Knockdown of long noncoding RNA HOXA-AS2 suppresses chemoresistance of acute myeloid leukemia via the miR-520c-3p/S100A4 axis. Cell Physiol Biochem. 2018;51:886–896.
- Lv J, Qiu M, Xia W, et al. High expression of long non-coding RNA SBF2-AS1 promotes proliferation in non-small cell lung cancer. J Exp Clin Cancer Res. 2016;35:75.
- Zhao QS, Li L, Zhang L, et al. Over-expression of lncRNA SBF2-AS1 is associated with advanced tumor progression and poor prognosis in patients with non-small cell lung cancer. Eur Rev Med Pharmacol Sci. 2016;20:3031–3034.
- Gao F, Feng J, Yao H, et al. LncRNA SBF2-AS1 promotes the progression of cervical cancer by regulating miR-361-5p/FOXM1 axis. Artif Cells Nanomed Biotechnol. 2019;47:776–782.
- Chen R, Xia W, Wang X, et al. Upregulated long non-coding RNA SBF2-AS1 promotes proliferation in esophageal squamous cell carcinoma. Oncol Lett. 2018;15:5071–5080.
- Zhang YT, Li BP, Zhang B, et al. LncRNA SBF2-AS1 promotes hepatocellular carcinoma metastasis by regulating EMT and predicts unfavorable prognosis. Eur Rev Med Pharmacol Sci. 2018;22:6333–6341.
- Zheng S, Jiang F, Ge D, et al. LncRNA SNHG3/miRNA-151a-3p/RAB22A axis regulates invasion and migration of osteosarcoma. Biomed Pharmacother. 2019;112:108695.
- Liu X, Lu X, Zhen F, et al. LINC00665 induces acquired resistance to gefitinib through recruiting EZH2 and activating PI3K/AKT pathway in NSCLC. Mol Ther Nucleic Acids 2019;16:155–161.
- Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet. 2016;17:272.
- Zhang H, Yang F, Chen SJ, et al. Upregulation of long non-coding RNA MALAT1 correlates with tumor progression and poor prognosis in clear cell renal cell carcinoma. Tumour Biol. 2015;36:2947–2955.
- Li P, Xue WJ, Feng Y, et al. Long non-coding RNA CASC2 suppresses the proliferation of gastric cancer cells by regulating the MAPK signaling pathway. Am J Transl Res. 2016;8:3522.
- Chen L, Hu N, Wang C, et al. Long non-coding RNA CCAT1 promotes multiple myeloma progression by acting as a molecular sponge of miR-181a-5p to modulate HOXA1 expression. Cell Cycle. 2018;17:319–329.
- Xu D, Yu J, Gao G, et al. LncRNA DANCR functions as a competing endogenous RNA to regulate RAB1A expression by sponging miR-634 in glioma. Biosci Rep. 2018;38:BSR20171664.
- Gao X, Zhao S, Yang X, et al. Long non-coding RNA FLVCR1-AS1 contributes to the proliferation and invasion of lung cancer by sponging miR-573 to upregulate the expression of E2F transcription factor 3. Biochem Biophys Res Commun. 2018;505:931–938.
- Fang F, Chang R, Yu L, et al. MicroRNA-188-5p suppresses tumor cell proliferation and metastasis by directly targeting FGF5 in hepatocellular carcinoma. J Hepatol. 2015;63:874–885.
- Peng Y, Shen X, Jiang H, et al. MiR-188-5p suppresses gastric cancer cell proliferation and invasion via targeting ZFP91. Oncol Res. 2018;27:65–71.
- Zhang H, Qi S, Zhang T, et al. miR-188-5p inhibits tumour growth and metastasis in prostate cancer by repressing LAPTM4B expression. Oncotarget 2015;6:6092.
- Reddy KB. MicroRNA (miRNA) in cancer. Cancer Cell Int. 2015;15:38.
- Saotome Y, Winter CG, Hirsh D. A widely expressed novel C2H2 zinc-finger protein with multiple consensus phosphorylation sites is conserved in mouse and man. Gene 1995;152:233–238.
- Tompkins V, Hagen J, Zediak VP, et al. Identification of novel ARF binding proteins by two-hybrid screening. Cell Cycle. 2006;5:642–647.
- Jin HR, Jin X, Lee JJ. Zinc-finger protein 91 plays a key role in LIGHT-induced activation of non-canonical NF-κB pathway. Biochem Biophys Res Commun. 2010;400:581–586.
- Ma J, Mi C, Wang KS, et al. Zinc finger protein 91 (ZFP91) activates HIF-1α via NF-κB/p65 to promote proliferation and tumorigenesis of colon cancer. Oncotarget. 2016;7:36551.
- Unoki M, Okutsu J, Nakamura Y. Identification of a novel human gene, ZFP91, involved in acute myelogenous leukemia. Int J Oncol. 2003;22:1217–1223.
