Abstract
Background
The purpose of our research was to explore potential value of lncRNA-BC050642 in osteosarcoma origination and prognosis.
Methods
In this study, the tissue specimens were collected from 97 osteosarcoma patients and 97 age-matched healthy controls. Besides, human osteosarcoma cell lines U2OS and normal osteoblastic cell line hFOB1.19 were selected for experiments in vitro. lncRNA-BC050642 levels were measured through quantitative real-time polymerase chain reaction (qRT-PCR). Relative expression of the gene c-myc was determined via ELISA analysis. x2 test was implemented to appraise possible relationship between BC050642 level and clinicopathological features. Cell proliferation assay, plate colony formation assay, and cell apoptosis assay were adopted to analyze the influence of BC050642 on tumor development. Besides, prognostic value of BC050642 was estimated using Kaplan–Meier and cox regression analysis.
Results
BC050642 levels showed distinctive increases in osteosarcoma tissues and cell lines compared with controls. And c-myc expression was down-regulated in osteosarcoma. There was a negative correlation between the expressions of BC050642 and c-myc. BC050642 expression was proved to be significantly correlated with Ennking and histological type. Up-regulated BC050642 could promote cell proliferation, induce colony formation and meanwhile inhibit cell apoptosis.
Conclusions
BC050642 is up-regulated in osteosarcma and its over-expression promotes tumor development via down-regulating the expression of c-myc. It also may be an independent prognostic biomarker for osteosarcoma.
Introduction
Osteosarcoma, a most common primary malignancy in bone, represents a leading cause of cancer-related death among adolescents and young adults [Citation1]. Despite its relatively low incidence, osteosarcoma shows soaring malignancy because of its high propensity of metastasis and local relapse [Citation2]. Main treatments for osteosarcoma are surgical excision and chemotherapy. But, it is frequent for the patients to develop lung metastasize which contributes to the cases’ deaths for the most part, and curative rate of osteosarcoma is low, with a 5-year survival rate of < 30% [Citation3]. Recent years, comprehensive therapies make 5 years survival rate raise to 60–70% [Citation4]. Whereas, main mechanism of osteosarcoma occurrence and development remain to be explored. Moreover, osteosarcoma still shows poor prognosis and serious harm. Therefore, it is necessary to explore its mechanisms and to develop schemes for the improvement of its diagnosis, treatment and prognosis.
Long non-coding RNAs (lncRNAs) refer to transcribed RNA molecules possessing a length of over 200 nt, with limited or no protein coding ability [Citation5,Citation6]. LncRNAs can regulate gene expression at epigenetic, transcriptional, and post-transcriptional levels [Citation7]. It is reported that lncRNAs play an important role in many cell processes such as proliferation, apoptosis, cell migration, and tumor metastases [Citation8–11]. lncRNAs could function as oncogene or tumor suppressor in different cancers [Citation12–16]. Linc0974 as a new biomarker interacts with KRT19 to promote proliferation and metastasis of hepatocellular carcinoma [Citation17]. lncRNA THOR can directly target the middle region of SOX9 3′-UTR, and then increase osteosarcoma cell stemness and migration by enhancing SOX9 mRNA stability [Citation18]. LINC00963 can promote the proliferation and invasion of osteosarcoma by inhibiting miR-204-3p/FN1 axis, and is a potential therapeutic target for osteosarcoma [Citation19]. What’s more, LncRNA BC050642 was found to be associated with c-myc promoter-binding protein-1, while the expression of c-myc protein was correlated with osteosarcoma prognosis [Citation20]. Therefore, we hypothesized that lncRNA-BC050642 might be associated with progression of osteosarcoma. However, the related studies had been rarely reported.
In our study, the expressions of BC050642 and c-myc in osteosarcoma were detected by qRT-PCR and ELISA, respectively. The role of BC050642 in the progress of osteosarcoma, including in cell proliferation, colony formation and cell apoptosis, was explored. The relationships of BC050642 with clinicopathologic characteristics and overall survival were analyzed to determine prognostic value of BC050642.
Material and methods
Patients and specimens
Ninety-seven osteosarcoma patients (29 females and 68 males, with a median age of 30.09 ± 11.54 years) were selected in our study. The patients were histopathologically confirmed independently by two pathologists. If there was disagreement, the histological results would be confirmed by the superior physician. All of the patients had undergone no treatments before operation. Besides, 97 healthy people with matched age were taken as controls. The study was approved by the Ethics Committee of Academy of Orthopedics of Guangdong Province, the Third Affiliated Hospital, Southern Medical University in Guangzhou province (approval no. 2017–0120). All patients signed written informed consents in advance.
Osteosarcoma tissues, paired adjacent tissues and healthy tissues were collected and frozen in liquid nitrogen immediately, and then stored at −80 °C for RNA extraction. Clinicopathologic profiles such as age, sex, tumor site, Ennking, Histological type, therapies, distant metastasis and recurrence were documented in a database. Follow-up was operated for 5 years and subjects dying from unexpected events or other illnesses were removed from this research.
Cell culture and cell transfection
Human osteosarcoma cell lines U2OS and normal osteoblastic cell line hFOB1.19 were gained from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). U2OS cell lines were maintained in Dulbecco’s modified Eagle’s medium while hFOB1.19 in DMEM/F-12 (1:1; HyClone, Logan, UT). All mediums were supplemented with 10% fetal bovine serum (FBS; Gibco, NY), 1% penicillin/streptomycin and 2 mM glutamine, and incubated at 37 °C in 5% CO2-humidified atmosphere.
U2OS cell lines were seeded in 96-well plates (6 × 103 cells/well) until the concentration reached to 50%. The cells were transfected with siRNA targeting BC050642 or with non-specific control siRNA (si-NC), using the Lipofectamine 2000 (Invitrogen, Carlsbad, CA) abide by relevant guidance overnight. All experiments were repeated three times.
RNA extraction and qRT-PCR analysis
Total RNA was extracted from tissue samples and cell lines adopting TRIzol (Invitrogen). RT-PCR reaction was conducted in the Applied Biosystems 7900 Fast Real-Time PCR system (Applied Biosystems, Foster City, CA) after synthesizing the first chain of cDNA with TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems). U6 was chosen as endogenous control for BC050642. Relative quantification of BC050642 was calculated via comparative cycle threshold (CT) method. All samples were in triplicate.
Plasmid construction and oligonucleotides
In the study, we constructed the following vectors: pcDNA3.1/c-myc. The sequences of primers were: forward-5′-CCGGGAATTCCTGGATTTTTTTCGGGTAGTG-3′ and reverse-5′-CCGGCTCGAGTTACGCACAAGAGTTCCGTAG-3′.
ELISA analysis
Total protein was isolated from tissue samples and cell lines. Then ELISA kit (R&D systems, Minneapolis, MN and Biocheck Inc., Foster City, CA, respectively) was used to measure the expression of the protein encoded by c-myc.
Cell proliferation assay
Osteosarcoma cell lines U2OS and normal osteoblastic cell line hFOB1.19 transfected with siRNAs or si-NC were seeded in 96-well plates (2 × 104/well), and cell viability at different time points (0, 24, 48 and 72 h) was determined at 450 nm adopting an enzyme immunoassay analyzer (Bio-Rad, Hercules, CA). Every trial was rerun thrice.
Plate colony formation assay
Logarithmic growth phase cells were collected, and adjusted to the density of 1 × 103/ml. The cells were seeded in 96-well plates. Medium was changed every 3 days, for 14 days of incubation. The cells were stained using crystal violet and then counted. Colony formation was quantified using colony formation number.
Cell apoptosis assay
Annexin V Apoptosis Detection Kit APC (eBioscience, San Diego, CA) was used to analyze cell apoptosis. First, logarithmic phase cells were selected applying three compound perforations, and adjusted to the density of 1 × 106/ml. Then, they were rinsed utilizing binding buffer, and staining buffer was supplemented to resuspend precipitate. Later, 100 μl cell suspension was employed for dyeing via 5 ml Annexin V-APC. Following 15 min of incubation in dark, the mixture was tested operating FACS Calibur (BD Biosciences, Franklin Lakes, NJ).
Statistical analysis
All quantified data were introduced as mean ± standard deviation (SD). Shapiro–Wilk and Levene tests were used to detect the normality and homogeneity of variance of the continuous variables. If the normality and homogeneity of variance were satisfied, Student’s t-test was performed for their comparison between two groups while one-way analysis of variance (ANOVA) analysis was applied for the analyses among three groups, otherwise, non-parametric test (Mann–Whitney U-test) was used. Chi-square test was conducted to assess possible link of clinicopathologic characteristics with BC050642 expression. The survival curves of the included patients based on their expression of BC050642 were plotted using Kaplan–Meier method with log rank test. In addition, cox regression analysis was performed to estimate the clinical significance of clinical parameters and BC050642 expression for the patients with osteosarcoma. Difference had statistical significance if p < .05. Statistical analysis was performed using SPSS version 13.0 software (SPSS Inc., Chicago, IL).
Result
BC050642 expression was increased in osteosarcoma tissues and cell lines
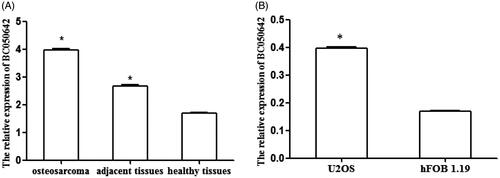
In this study, we detected BC050642 levels in osteosarcoma, adjacent and healthy tissues as well as in tumor cell lines U2OS and normal osteoblastic cell line hFOB1.19 through qRT-PCR. All the continuous data obtained in our study conformed to the normality and homogeneity of variance. Student’s t-test results showed that BC050642 expression was significantly higher in osteosarcoma tissues than in adjacent and healthy controls (, p < .001). Compared with normal osteoblastic cell line hFOB1.19, BC050642 expression was significantly up-regulated in osteosarcoma cell lines U2OS (, p < .001). Taken together, these results confirmed that BC050642 expression showed increased tendency in osteosarcoma.
Figure 1. The expression of BC050642 in osteosarcoma tissues and cell lines detected using qRT-PCR method. (A) The expression of BC050642 was higher in osteosarcoma tissues than in adjacent tissues and healthy W tissues. Asterisk (*) compared to healthy tissues, p < .05. (B) The expression of BC050642 was increased in osteosarcoma cell lines U2OS compared to normal osteoblastic cell line hFOB1.19. Asterisk (*) compared to hFOB1.19 cells, p < .05.
The expression of c-myc in osteosarcoma patients
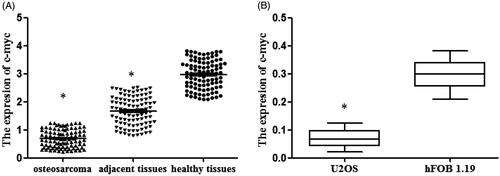
C-myc was considered to be related to the expression of BC050642 and to the prognosis of osteosarcoma, we measured its expression using ELISA analysis. The outcome proved that c-myc expression was decreased in cancer tissues compared to adjacent and healthy ones (, p < .05). As for in cell lines, c-myc expression was also lower in U2OS than in hFOB1.19 (, p < .05). These findings suggested that the expressions of C-myc were decreased in osteosarcoma.
Figure 2. The expression of c-myc in osteosarcoma tissues and cell lines detected by the method of ELISA. (A) The levels of c-myc in osteosarcoma tissues, adjacent tissues and healthy tissues. Asterisk (*) compared to healthy tissues, p < .05. (B) Osteosarcoma cell lines U2OS and normal osteoblastic cell line hFOB1.19 were collected. The expression of c-myc was analyzed by RT-qPCR. Asterisk (*) compared tohFOB1.19 cells, p < .05.
BC050642 promoted osteosarcoma cell proliferation, increased colony formation and inhibited cell apoptosis
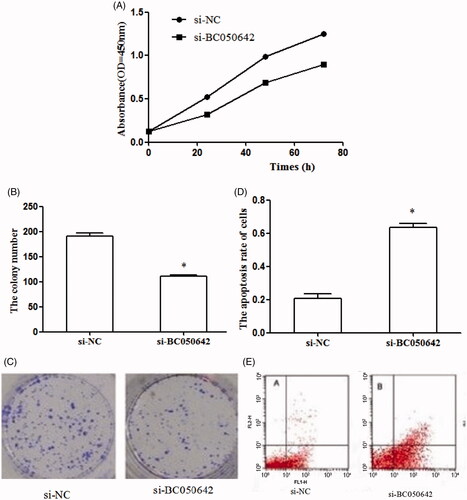
To determine whether BC050642 functioned as an oncogene in osteosarcoma, we conducted MTT assay, colony formation assay and FACS assay. MTT assay showed that the proliferation of cells transfected with si-BC050642 was significantly decreased than those transfected with si-NC (). The clone number of cells transfected with si-BC050642 was reduced when compared with those transfected with si-NC (, p < .05). Besides, apoptosis percentage of U2OS cells was significantly increased in si-BC050642 group compared to si-NC group (, p < .05). These findings suggested that the knockdown of BC050642 resulted in inhibition on cell proliferation and colony formation, and promoting effects on cell apoptosis of osteosarcoma cell lines U2OS, revealing the oncogenic potential of BC050642 in osteosarcoma.
Figure 3. The effects of BC050642 on biological behaviors of osteosarcoma cell line U2OS. (A) The down-regulation of BC050642 inhibited cell proliferation; (B, C) the down-regulation of BC050642 reduced colony formation of osteosarcoma cells, asterisk (*) compared to Si-NC, p < .05; (D, E) the down-regulation of BC050642 induced cell apoptosis, asterisk (*) compared to Si-NC, p < .05.
C-myc-inhibited osteosarcoma cell proliferation, reduced colony formation and induced cell apoptosis
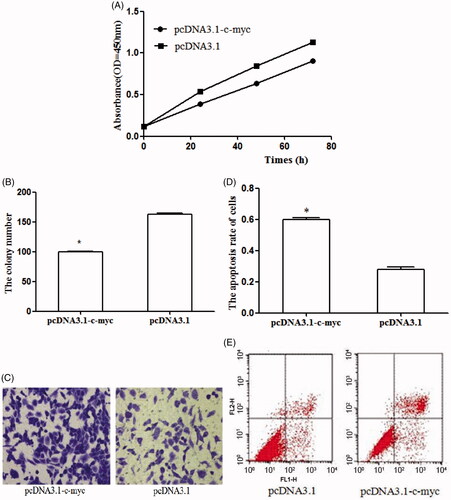
To examine the effect of c-myc on osteosarcoma, pcDNA 3.1-c-myc was designed to enhance the expression of c-myc in osteosarcoma cell lines. The efficiency of the transfection was detected through qRT-PCR. As displayed in , the transfection of pcDNA 3.1-c-myc obviously promote the expression of c-myc in osteosarcoma cells. Following, the cell experiments were designed to explore the function of c-myc in osteosarcoma. The restored expression of c-myc inhibited cell proliferation () and suppressed colony formation number (). In addition, cell apoptosis assay showed that the over-expression of c-myc promoted cell apoptosis (). These results demonstrated that the anti-tumor of c-myc on cell proliferation, colony formation and apoptosis.
Figure 4. C-myc expression was significantly higher in cells transfected with pcDNA3.1-c-myc than those transfected with empty vector (pcDNA3.1) (*p < .05 represented the significant difference between the compared two).
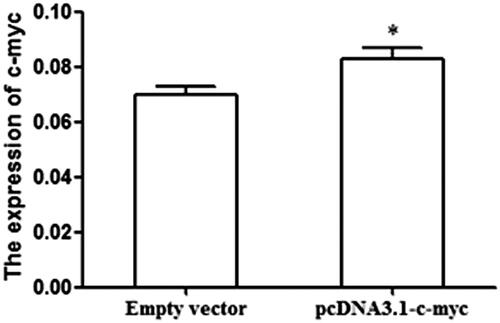
Figure 5. The effects of c-myc on biological behaviors of osteosarcoma cell line U2OS. (A) The up-regulation of BC050642 suppressed cell proliferation; (B, C) representative colony formation assay images of U2OS cells transfected by pcDNA3.1-vector and pcDNA3.1-c-myc. The numbers of the colonies in vector-transfected controls reached 100%, asterisk (*) compared to pcDNA3.1-vector, p < .05; (D, E) C-myc over-expression promoted cell apoptosis, asterisk (*) compared to pcDNA3.1-vector, p < .05.
The relationship between clinicopathologic characteristics and BC050642
To investigate whether the expression of BC050642 was related to the progression of osteosarcoma, we estimated its relationship with clinicopathologic characteristics. According to the expression of BC050642, osteosarcoma cases were sorted into high and low BC050642 expression groups, based on a median expression of 3.958 ± 0.504. As displayed in , Ennking (p = .000) and histological type (p=.022) were significantly related to the expression of BC050642. Whereas, BC050642 had no relationship with age, gender, tumor site, therapies, distant metastasis or recurrence.
Table 1. The relationship between clinicopathologic characteristics and BC050642 in patients with osteosarcoma.
Prognostic value for BC050642 in osteosarcoma
Kaplan–Meier analysis revealed shorter overall survival for the subjects harboring high BC050642 levels when compared with low ones (log-rank test, p < .001, ). Cox regression analysis was used to further inspect the clinical significance of BC050642 in the cancer prognosis. As a result, Ennking (HR = 1.535, 95% CI=0.355–6.641, p = .012), distant metastasis (HR = 4.241, 95% CI = 1.431–12.564, p = .009) and BC050642 (HR = 13.846, 95% CI = 2.362–91.153, p = .004) acted as influential factors for osteosarcoma prognosis (). Furthermore, BC050642 might be an independent biomarker for cancer prognosis.
Figure 6. Kaplan–Meier analysis for osteosarcoma patients according to their expression of BC050642. Patients with high BC050642 expression had shorter overall survival than those with low BC050642 expression (log-rank test, p<.001).
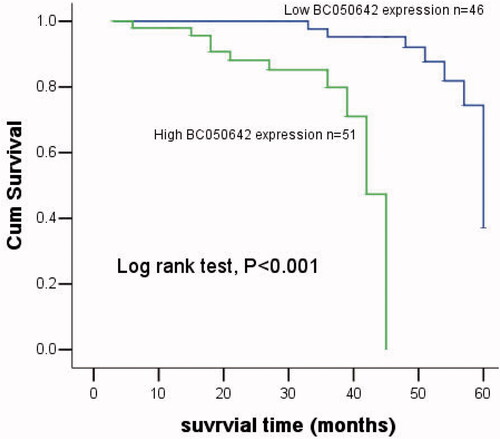
Table 2. Multivariate analysis adjusting clinical factors for prognostic value of BC050642 in patients with osteosarcoma.
Discussion
Osteosarcoma stands for a differentiation disease which was caused by genetic changes that interrupt osteoblast differentiation from mesenchymal stem cells [Citation21]. It generally stems from the metaphysis of long bones in children and juveniles, such as distal femur, proximal tibia and proximal humerus [Citation22]. Cure rate is < 65% for localized osteosarcoma patients, while cases showing metastasis when diagnosed frequently experience poorer prognosis [Citation23]. Despite great advancements in osteosarcoma explorations [Citation24], its exact mechanism is beyond total comprehension.
LncRNAs have been recently considered to be oncogene or tumor suppressor in different cancers. Without protein-coding capacity, they can regulate gene expression and is involved in the development of human diseases, including tumors [Citation25,Citation26]. More and more lncRNAs have been confirmed to act as diagnostic and prognostic markers or regulators in tumor development. However, studies about lncRNAs in osteosarcoma were rare. Ivan Pasic et al. [Citation27], found the deletion of lncRNA-LOC285194 and BC040587 might be a reason of osteosarcoma onset, and predicted poor prognosis, via exploring their functions in vitro. Zhang et al. [Citation28] reported that lncRNA-TUG1 could not only inhibit cell proliferation but promote apoptosis, which might provide a therapeutic strategy for osteosarcoma. LncRNA loc285194 was considered as a p-53 regulated tumor suppressor and inhibited tumor cell growth in osteosarcoma both in vitro and in vivo [Citation29]. Reportedly, 25,733 lncRNAs including 403 lncRNAs over-regulated and 798 lncRNA under-regulated have been discovered in osteosarcoma by Li et al. [Citation30]. In our study, we detected BC050642 levels in osteosarcoma tissues and cell lines. The results revealed that the expression of BC050642 in osteosarcoma tissues or cell lines was higher than that in controls. The analysis of the relationship between clinicopathologic characteristics and BC050642 manifested that BC050642 was related to the progress of osteosarcoma. Biological functions of BC050642 in osteosarcoma cell lines were also studied in vitro. And, we accomplished MTT assay, colony formation assay and FACS assay. According to relevant results, the up-regulation of BC050642 could promote cell proliferation, increase colony formation and inhibit apoptosis in osteosarcoma. Taken together, BC050642 played a critical role in the development and progression of osteosarcoma, being an oncogene in the malignancy.
The c-myc gene, located on the long arm of chromosome-8 (8q24), is a member of myc family decreased in a various of cancers such as lung cancer, breast cancer, prostate cancer, leukemias and lymphomas [Citation31,Citation32]. C-myc can not only induce histone acetyltransferase (HAT) activity but also promote RNA polymerase II (RNAPII) clearance [Citation33]. C-myc has been reported to be involved in several cancers through interacting with lncRNAs. For instance, c-myc increased the expression of lncRNA-CCAT1 and promoted cell proliferation in gastric carcinoma as well as in colon cancer [Citation34,Citation35]. Yang et al. [Citation36] found lncRNA-GHET1 can regulate cell proliferation via increasing c-myc mRNA stability and expression while Zhang et al. [Citation37] insisted the over-expression of lncRNA-H19 induced by c-myc could regulate cell proliferation and predict poor prognosis in gastric cancer. C-myc translation was controlled by lncRNA-GAS5 and the interaction between them and eIF4E was important in translation regulation in lymphoma [Citation38]. Liao et al. [Citation39] revealed critical role of lncRNA-XLOC-010588 expression in cell proliferation via decreasing c-myc expression. Recently, c-myc was related to the prognosis of osteosarcoma as well as to the over-expression of BC050642 [Citation40]. Therefore, we transfected c-myc overexpression vector pcDNA 3.1-c-myc into osteosarcoma cell lines and investigated the effects of c-myc on osteosarcoma development. The result indicated that c-myc over-expression could inhibit cell proliferation, decrease colony formation and induce cell apoptosis.
To investigate BC050642 merit in osteosarcoma prognosis, we conducted Kaplan–Meier and cox regression analysis. Kaplan–Meier showed osteosarcoma cases enjoying low BC050642 levels lived longer than high subjects. Besides, cox regression analysis demonstrated that high BC050642 expression as well as Ennking and distant metastasis significantly influenced osteosarcoma outcomes, and they had the potential of independently predicting the patients’ prognosis.
Conclusion
In conclusion, the expression of BC050642 is increased in osteosarcoma and closely correlated with Ennking and histological type. The over-expression of BC050642 promotes cell proliferation, induces colony formation and inhibits cell apoptosis via down-regulating the expression of c-myc. High BC050642 may predict dismal clinical outcomes for the patients with osteosarcoma.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Tsai HC, Tzeng HE, Huang CY, et al. WISP-1 positively regulates angiogenesis by controlling VEGF-A expression in human osteosarcoma. Cell Death Dis. 2017;8:e2750.
- Kimura Y, Tomihara K, Tachinami H, et al. Conventional osteosarcoma of the mandible successfully treated with radical surgery and adjuvant chemotherapy after responding poorly to neoadjuvant chemotherapy: a case report. J Med Case Rep. 2017;11:210.
- Haghiralsadat F, Amoabediny G, Naderinezhad S, et al. Codelivery of doxorubicin and JIP1 siRNA with novel EphA2-targeted PEGylated cationic nanoliposomes to overcome osteosarcoma multidrug resistance. Int J Nanomed. 2018;13:3853–3866.
- Magalhães M, Almeida M, Tavares-da-Silva E, et al. miR-145-loaded micelleplexes as a novel therapeutic strategy to inhibit proliferation and migration of osteosarcoma cells. Eur J Pharm Sci. 2018;123:28–42.
- Dragomir M, Chen B, Calin GA. Exosomal lncRNAs as new players in cell-to-cell communication. Transl Cancer Res. 2018;7:S243–S252.
- Deng F, Zhang X, Wang W, et al. Identification of Gossypium hirsutum long non-coding RNAs (lncRNAs) under salt stress. BMC Plant Biol. 2018;18:23.
- Rathinasamy B, Velmurugan BK. Role of lncRNAs in the cancer development and progression and their regulation by various phytochemicals. Biomed Pharmacother. 2018;102:242–248.
- Song X, Luo X, Gao Q, et al. Dysregulation of LncRNAs in placenta and pathogenesis of preeclampsia. Curr Drug Targets. 2017;18:1165–1170.
- Gioia R, Drouin S, Ouimet M, et al. LncRNAs downregulated in childhood acute lymphoblastic leukemia modulate apoptosis, cell migration, and DNA damage response. Oncotarget. 2017;8:80645–80650.
- Ma SC, Li Q, Peng JY, et al. CLDN5 affects lncRNAs acting as ceRNA dynamics contributing to regulating blood brain barrier permeability in tumor brain metastasis. Oncol Rep. 2018;39:1441–1453.
- Duguang L, Jin H, Xiaowei Q, et al. The involvement of lncRNAs in the development and progression of pancreatic cancer. Cancer Biol Ther. 2017;18:927–936.
- Xie J, Lin D, Lee DH, et al. Copy number analysis identifies tumor suppressive lncRNAs in human osteosarcoma. Int J Oncol. 2017;50:863–872.
- Grossi E, Sánchez Y, Huarte M. Expanding the p53 regulatory network: LncRNAs take up the challenge. Biochim Biophys Acta. 2016;1859:200–208.
- Wan X, Ding X, Chen S, et al. The functional sites of miRNAs and lncRNAs in gastric carcinogenesis. Tumor Biol. 2015;36:521–532.
- Xiong Y, Wang T, Wang M, et al. Long non-coding RNAs function as novel predictors and targets of non-small cell lung cancer: a systematic review and meta-analysis. Oncotarget. 2018;9:11377–11386.
- Wu J, Hann SS. Functions and roles of long-non-coding RNAs in human nasopharyngeal carcinoma. Cell Physiol Biochem. 2018;45:1191–1204.
- Tang J, Zhuo H, Zhang X, et al. A novel biomarker Linc00974 interacting with KRT19 promotes proliferation and metastasis in hepatocellular carcinoma. Cell Death Dis. 2014;5:e1549.
- Wu H, He Y, Chen H, et al. LncRNA THOR increases osteosarcoma cell stemness and migration by enhancing SOX9 mRNA stability. FEBS Open Bio. 2019;9:781–790.
- Zhou Y, Yin L, Li H, et al. The LncRNA LINC00963 facilitates osteosarcoma proliferation and invasion by suppressing miR-204-3p/FN1 axis. Cancer Biol Ther. 2019;12:1–8.
- Dong Y, Liang G, Yuan B, et al. MALAT1 promotes the proliferation and metastasis of osteosarcoma cells by activating the PI3K/Akt pathway. Tumour Biol. 2015;36:1477–1486.
- Ren T, Piperdi S, Koirala P, et al. CD49b inhibits osteogenic differentiation and plays an important role in osteosarcoma progression. Oncotarget. 2017;8:87848–87859.
- Krishnamurthy A, Arulmolichelvan A. The management challenges in an unusual case of primary osteosarcoma of the rib in an adult patient. Indian J Surg. 2017;79:363–366.
- Coventon J. A review of the mechanism of action and clinical applications of sorafenib in advanced osteosarcoma. J Bone Oncol. 2017;8:4–7.
- Park SH, Lee J, Kang MA, et al. Mitoxantrone induces apoptosis in osteosarcoma cells through regulation of the Akt/FOXO3 pathway. Oncol Lett. 2018;15:9687–9696.
- Zou H, Wu LX, Yang Y, et al. lncRNAs PVT1 and HAR1A are prognosis biomarkers and indicate therapy outcome for diffuse glioma patients. Oncotarget. 2017;8:78767–78780.
- Li S, Chen X, Liu X, et al. Complex integrated analysis of lncRNAs-miRNAs-mRNAs in oral squamous cell carcinoma. Oral Oncol. 2017;73:1–9.
- Pasic I, Shlien A, Durbin AD, et al. Recurrent focal copy-number changes and loss of heterozygosity implicate two noncoding RNAs and one tumor suppressor gene at chromosome 3q13.31 in osteosarcoma. Cancer Res. 2010;70:160–171.
- Zhang Q, Geng PL, Yin P, et al. Down-regulation of long non-coding RNA TUG1 inhibits osteosarcoma cell proliferation and promotes apoptosis. Asian Pac J Cancer Prev. 2013;14:2311–2315.
- Liu Q, Huang J, Zhou N, et al. LncRNA loc285194 is a p53-regulated tumor suppressor. Nucleic Acids Res. 2013;41:4976–4987.
- Li JP, Liu LH, Li J, et al. Microarray expression profile of long noncoding RNAs in human osteosarcoma. Biochem Biophys Res Commun. 2013;433:200–206.
- Kiaei A, Onsori H, Alijani A, et al. Detection of t(8;14) c-myc/IgH gene rearrangement by long-distance polymerase chain reaction in patients with diffuse large B-cell lymphoma. Hematol Oncol Stem Cell Ther. 2016;9:141–146.
- Melis MHM, Nevedomskaya E, van Burgsteden J, et al. The adaptive immune system promotes initiation of prostate carcinogenesis in a human c-Myc transgenic mouse model. Oncotarget 2017;8:93867–93877.
- Lombardi O, Varshney D, Phillips NM, et al. c-Myc deregulation induces mRNA capping enzyme dependency. Oncotarget. 2016;7:82273–82288.
- Yang F, Xue X, Bi J, et al. Long noncoding RNA CCAT1, which could be activated by c-Myc, promotes the progression of gastric carcinoma. J Cancer Res Clin Oncol. 2013;139:437–445.
- He X, Tan X, Wang X, et al. C-Myc-activated long noncoding RNA CCAT1 promotes colon cancer cell proliferation and invasion. Tumour Biol. 2014;35:12181–12188.
- Yang F, Xue X, Zheng L, et al. Long non-coding RNA GHET1 promotes gastric carcinoma cell proliferation by increasing c-Myc mRNA stability. FEBS J. 2014;281:802–813.
- Zhang EB, Han L, Yin DD, et al. c-Myc-induced, long, noncoding H19 affects cell proliferation and predicts a poor prognosis in patients with gastric cancer. Med Oncol. 2014;31:914.
- Hu G, Lou Z, Gupta M. The long non-coding RNA GAS5 cooperates with the eukaryotic translation initiation factor 4E to regulate c-Myc translation. PLoS One. 2014;9:e107016.
- Liao LM, Sun XY, Liu AW, et al. Low expression of long noncoding XLOC_010588 indicates a poor prognosis and promotes proliferation through upregulation of c-Myc in cervical cancer. Gynecol Oncol. 2014;133:616–623.
- Yan CH, Li F, Ma YC. Plumbagin shows anticancer activity in human osteosarcoma (MG-63) cells via the inhibition of S-Phase checkpoints and down-regulation of c-myc. Int J Clin Exp Med. 2015;8:14432–14439.
