Abstract
Objectives
MiR-34b is a tumour suppressor in different kinds of carcinomas. This study investigated the role of miR-34b in proliferation and apoptosis of cervical cancer.
Materials and methods
The expression of miR-34b in 60 cervical cancer patients were quantified by RT-PCR and correlated with their clinicopathological parameters. Besides, there is a significant reverse relationship between miR-43b and TGF-β1 expression in tumour tissues. Cell proliferation and apoptosis was detected by CCK-8 assays and flow cytometry in cell lines transfected with miR-34b mimics. Western blotting, quantitative reverse transcription-PCR (RT-PCR) and luciferase assays were conducted to analyze the regulation of TGF-β1 by miR-34b in cell lines.
Results
Here, we found expression of miR-34b to be downregulated in cervical cancer in comparison with the adjacent normal tissues. Expression levels of miR-34b were associated with enhanced malignant potential, such as tumour stage and stromal invasion. The overexpression of miR-34b potently suppressed cell proliferation and induced the apoptosis of cell lines.
Conclusions
MiR-34b and TGF-β1 contribute to cervical cancer cell proliferation and apoptosis and are potential targets for cervical cancer therapeutics.
Keywords:
Introduction
Cervical cancer is one of the most common cancers in women. Emerging evidence suggests cervical cancer is caused by genetic and epigenetic changes as well as different kinds of environmental factors [Citation1,Citation2]. Advances in cervical cancer therapy over the past decade have enhanced patient outcomes, and the 5-year survival rate of cervical cancer patients has been dramatically improved [Citation3]. However, the outcome remains poor and most of them died of long-term metastases eventually [Citation2]. Downregulation of certain tumour suppressor genes was confirmed to largely contribute to initiation, proliferation, invasion and metastasis of cervical cancer [Citation4]. Therefore, targeted gene therapy has been documented as a reasonable strategy for cervical cancer treatment [Citation5].
MicroRNAs (miRNAs) are a group of small (∼22 nucleotides) endogenous non-coding RNAs and previous studies have demonstrated their posttranscriptional regulation effects through RNA interference [Citation6,Citation7]. MiRNAs have been reported to be involved in a series of biological processes and demonstrate impacts on many diseases including cancer [Citation8,Citation9]. Abnormal miRNAs expressions have been reported in different types of cancers. These miRNAs might demonstrate either diagnostic or prognostic effects and have important impacts in altering diseases phenotypes through targeting biological functions in tumourigenesis. In a most recent study, it was showed that eight microRNAs (miR-9-5p, miR-136-5p, miR-148a-3p, miR-190a-5p, miR-199b-5p, miR-382-5p, miR-597-5p and miR-655-3p) were expressed differently in the HPV16-positive cervical cancer group with the hybridization method, and HPV16-positive normal group [Citation10]. In another review, recent data on the role of microRNAs in the metastasis of HPV-related cancers and on their possible clinical relevance as biomarkers of metastatic disease and/or as therapeutic targets is presented [Citation11].
Several studies have demonstrated that miR-34b was reduced or lost in a number of cancers, including breast cancer, epithelial ovarian cancer and Gastric Cancer [Citation12–14]. The association between a polymorphism in miR-34b and risk of cancer was also reported in independent studies and meta-analyses [Citation15,Citation16]. There were also some studies focusing on the effect of miR-34b in the development of cervical cancer. In a study including 88 cervical cancer cases, it was reported that miR-34b-3p were found to be frequently downregulated in cancers suggesting miRNA-mediated deregulation of APOBEC3A expression in cancer patients harbouring this particular deletion polymorphism [Citation17]. MiR-34b was also reported to distinguish the high-grade CIN specimens from normal cervical epithelium [Citation18]. However, a fundamental question was whether miR-34b has therapeutic potential for cervical cancer. In addition, the signalling mechanisms by which miR-34b functions remain to be determined. Thus we conducted this study to detect the expression pattern of miR-34b in cervical cancers and demonstrate the regulation effect of miR-34b through in-vitro studies.
Materials and methods
Clinical specimen preparation
A total of 60 clinical cervical cancer tissue specimens were collected from the Zhongnan Hospital of Wuhan University. The cervical cancer samples were originally from patients between 27 and 70 years old. Exclusive criteria: individuals with diabetes, other malignancy or severe diseases. Each patient provided informed consent and the study was approved by the Ethics Committee of Zhongnan Hospital of Wuhan University. All tissue samples were snap-frozen in liquid nitrogen and saved at −80 °C before use.
RNA extraction and miRNA detection
The total RNA was extracted using TRIzol reagent (Invitrogen: Thermo Fisher Scientific, Inc.). The miRNA samples were extracted from cervical cancer tissues and cell lines with RNeasy Mini Kits (Qiagen, CA, USA) and all the procedures were conducted according to the manufacturer’s manual. Concentration and quality of extracted RNA were assessed through measuring absorbance at 260/280 nm with BioPhotometer (Eppendorf, Germany). The miR-34b expression level was determined by real-time PCR with TaqMan MicroRNA Assay Kits (ABI, Foster City, CA, USA). The primers used in this study were the following: miR-34b: forward 5′-CCAGGACCAGAGGAAACCT-3′, reverse 5′-GCTAGCCTCTGGATTTGA-3′. GAPDH: forward 5′-ATGTCGTGGAGTCTACTGGC-3′, reverse 5′-TGACCTTGCCCACAGCCTTG-3′. TGF1β mRNA expression was detected with SYGR green real-time PCR (TAKARA, Tokyo Japan). The real-time PCR data were normalized with 2−ΔΔCt method relative to GAPDH.
Cell culture
Immortalized human HaCaT keratinocytes and cervical cancer cell line C33a were purchased from China Center For Type Culture Collection (Wuhan, China). Both cell lines were maintained in Minimum Essential Medium (MEM, Thermo Fisher Scientific, Rockford, IL, USA) supplemented with 10% FBS (Thermo Fisher Scientific). HaCaT cells were grown in Defined Keratinocyte-SFM medium (Thermo Fisher Scientific). All cells were cultured in a humidified incubator with 5% CO2 at 37 °C.
Cell viability assay
Cell Counting Kit-8 (CCK-8) was used in the qualitative detection of cell viability in this study. The CCK-8 method was based on the conversion of a water-soluble tetrazolium WST-8, to a water-soluble formazan dye upon reduction by dehydrogenases in the presence of an electron carrier. All the experiments were conducted according to the manufacturer’s protocol. Generally speaking, the cells were plated in 96-well plates, then 10 µL of CCK-8 solution was added to each well, and the samples were incubated for one hour before the absorbance was measured at 450 nm. Each experiment was conducted for three times.
Wound healing assay
The cultured cells will be harvested and subjected to a wound healing assay. Cells in all the groups were seeded in 6-well plates at 5 × 105 cells/well and cultured for 12 h until confluent. Then, the plates in all the groups were washed twice with PBS and culture medium was changed to medium with 5% FBS for 24 h. Scratch wounds were made across the center of all the wells using 200 μL pipette tips. Wound images after 36 h treatment of miR-34a or miR-control mimics transfection were captured with an inverted microscope. Image J was used to detect the wound healing rate and each experiment was conducted for three times.
Apoptosis assay
In this study, flow cytometery with Annexin V and PI stain was used in the detection of apoptosis. The cellular samples were harvested at 72 h post-transfection by trypsinization and single cell samples were used in the assay. In the assay, cells samples were at a density of 1 × 106 cells/mL. After double staining with Annexin V/Alexa Fluor 647 and propidium iodide (PI) for 15 min at room temperature with the Annexin V-Alexa Apoptosis Detection Kit (Fcmacs, China). After that, samples were analyzed on a FACScan flow cytometer equipped with Cell Quest software (BD Biosciences) and the results were used in the apoptotic rate analyses.
Western blot analysis
The protein samples were extracted from each group with RIPA lysis buffer. After centrifuging at 15,000 × g for 20 min at 4 °C, the supernatants of homogenates were collected and protein quantification was conducted using the BCA method. A total of 50 μg of protein was used in each sample and separated 10% SDS-polyacrylamide gel electrophoresis and transferred. After running the gels, the protein binds was transformed to PVDF membranes and after that, the membranes were washed with Tris-buffered saline (TBS) and then blocked with 5% non-fat dry milk in Tris-buffered saline Tween-20 (TBST) for 1 h, and then incubated with primary antibody. After washing with TBST twice, a secondary antibody conjugated to horseradish peroxidase for 1 h at room temperature. The protein expression level was normalized to the corresponding GAPDH in this study.
Luciferase activity assay
To detect and verify the target gene of miR-34b, luciferase activity assay was obtained in this study. Briefly, the cultured cells were seeded in a 12-well plate in the density of 1 × 105 cells/well. After co-transfecting with wild type or mutated 3’-UTRs of TGF-β1 luciferase reporter constructs, either miR-34b or control mimic was transfected in both groups were conducted with Lipofectamine 3000 according to supplier’s protocol. The cell was then harvested after treatment of 24 h and after that, luciferase intensity was examined by Dual-Luciferase Reporter Assay Kit (Promega, Wisconsin, WI, USA).
Statistical analysis
All the data were analyzed with SPSS 20.0 software (SPSS, Inc., Chicago, IL, USA). All the data were presented as mean ± SD and ANOVA or two-tail Student’s t-test was used to examine the statistical significance of comparison of the means in different groups. P < .05 was accepted as statistically significant.
Results
Association between miR-34b expression and clinicopathological parameters
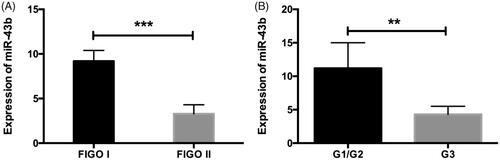
In order to determine the expression levels of miR-34b in cervical cancer tissues, quantitative RT-PCR detection was made in 60 cervical cancer tissues. We first evaluated the expression of miR-34b in cervical cancer tissue specimens. Our data showed that decreased miR-34b was associated with clinical pathological characteristics of cervical cancer cases. The results showed that the decreased expression of miRNA-34b expression in cervical cancer tissues was remarkably in high FIGO status and stages ().
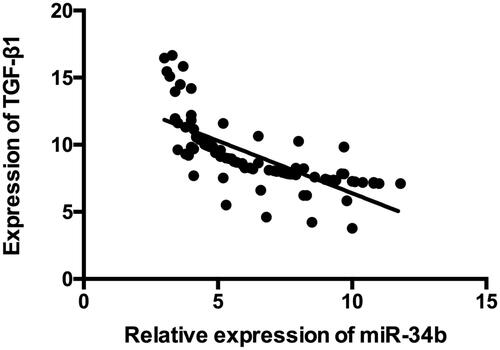
The analysis between miR-34b expression and clinicopathological parameters confirmed that less expression of miR-34b in cervical cancer cases was associated with larger tumour size, advanced FIGO stage and stromal invasion (P < .05). There was no significant difference in age, pathological type, tumour grade, lymph node metastasis or HPV infection (). We also detected the expressions of miR-34b and TGF-β1 in the cervical tissues. As showed in , it was found that miR-34b expression was inversely associated with the level of TGF-β1.
Table 1. Correlation of miRNA-34b expression with the clincopathological characteristics of the cervical cancer patients.
MiR-34b is decreased in cervical cancer cell line and influences the cell viability
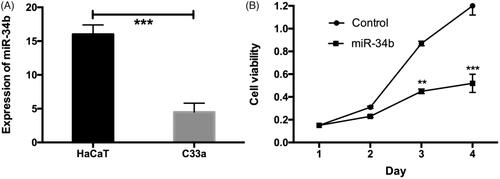
The expression of miRNA-34b was then measured in the cervical cancer cell line, C33a. The results indicated that, compared to the control group, miRNA-34b expression in C33a cells was significantly decreased (). Cervical cancer cell C33a was treated with 100 nM of control or miR-34b mimics for 24, 48, 72 h and measured the absorbance at 490 nm. The inhibition rate was calculated as following: inhibition rate (%) = (OD value of the control group − OD value of experimental group)/OD value of control group × 100%. Compared with the control group, the miR-34b treated group was inhibited in a dose and time-dependent manner. Cell proliferation was strongest inhibited when cells were treated with 100 nM miR-34b mimics for 72 h with the inhibition rate of 56.4% (P < .001) ().
Figure 3. MiR-34b was decreased in cervical cancer tissues and inhibited the cell viability. (A) MiR-34b was significantly decreased in cervical cancer cell line compared with that in the corresponding normal cell line. (B) MiR-214 significantly inhibited the cell viability in day 2 and 3. **P < .01; ***P < .001 compared with the control group.
MiR-34b induced apoptosis and inhibited C33a cell migration
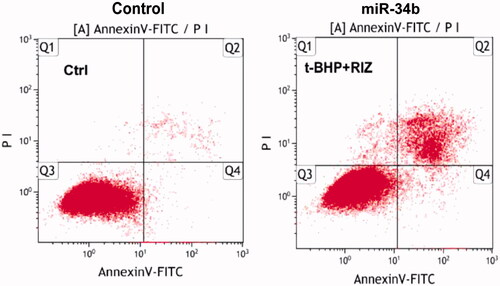
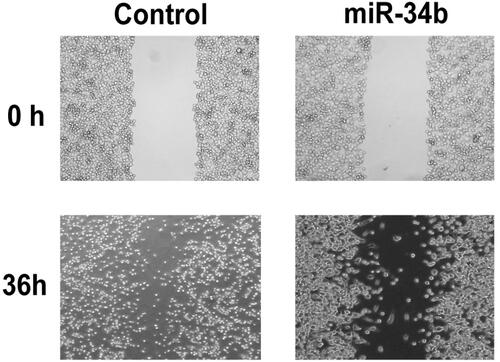
Flow cytometry was used to measure apoptosis after overexpression of miR-34b. As reported, the overexpression of miR-34b could induce apoptosis compared with negative controls in C33a cells (. Cell scratch assay showed that cell transfected with miR-34b mimics migrated slowly. Scratch in the control group was almost healed 36 h after the scratch had been made but not in miR-34b mimics group. These data showed that miR-34b inhibited C33a cell migration ().
MiR-34b directly targets TGF-β1 in cervical cancer cells
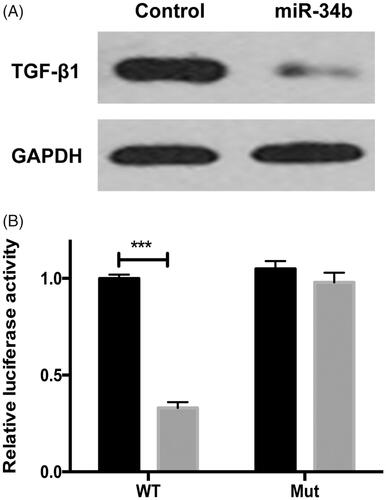
We used TargetScan 6.2 software to search for the potential target gene of miR-34b. TGF-β1was predicted to be a target of miR-34b. It was found that TGF-β1 expression was significantly decreased after the treatment of miR-34b (). Then, the luciferase activity assay showed that miR-34b significantly suppressed the WT 3’-UTR but not that of Mut 3’-UTR of TGF-1β luciferase activity in cervical cancer cells ().
Figure 6. TGF-β1 was a direct target of miR-34b. (A) Protein level of TGF-β1 and GAPDH was detected by Western blot in C33a cells transfected with miR-34b/ctrl. (B) C33a cells were co-transfected with miR-34a and WT or Mut 3’-UTR luciferase reporter construct. ***P < .001 compared with the control group.
Discussion
Emerging studies have revealed that miRNAs participate in the progression of various cancers including cervical cancer through regulation of expression of multiple target genes involved in the progression [Citation19]. Mounting pieces of evidence indicate that aberrant miRNA expression demonstrated an important regulatory effect in cancer development [Citation20]. Previous studies also demonstrated that the dysregulation of miRNAs potentially serves as a therapeutic target in various cancers including cervical cancer. Therefore, the study of the miRNA expression and functions in cervical cancer may help to improve the treatment strategy for refractory cervical cancer. In this study, we demonstrate that miR-34b expression is down-regulated in cervical cancer tissues. Forced overexpression of miR-214 is able to inhibit cell proliferation and induce apoptosis in C33a cells. Therefore, our study, for the first time, identifies that miR-34b might be a potential inhibitor in the progression of cervical cancer. However, further studies are still needed to investigate its roles in-vivo.
In this study, it was found that decreased miR-34b was associated with clinical pathological characteristics of cervical cancer cases. The results showed that the decreased expression of miRNA-34b expression in cervical cancer tissues was remarkably in high FIGO status and stages. This study was quite consistent with previous studies indicating that miR-34b could be a potential cancer suppressor of different cancers [Citation21,Citation22]. This result was clinically significant and demonstrated that miR-34b might be used for the following detection of its diagnostic or prognostic effect.
Previous evidence has suggested that EMT engages in primary tumour metastasis and reveals molecular mechanisms for cervical cancer metastasis [Citation23]. Importantly, TGF-β and TGF-β-related proteins have been confirmed as the major inducers of EMT event in cancer development, and the convergence of TGF-β signaling pathways is essential for EMT. EMT program was triggered by the exposition of both epithelial cell lines to TGF-β1. Based on these findings, it can be seen that HOXC6 gene silencing inhibits the EMT in cervical cancer by inactivating the TGF-β/smad signaling pathway [Citation24]. In addition, at the molecular level, our results reveal that TGF-β1 is a direct target of miR-34b in cervical cancer cells. As reported in a previous study, TGF-β1 pathway was reported to be a key regulation pathway for cervical cancer [Citation25]. These studies revealed the requirement of TGF-β1 for cervical cancer development. Up-regulation of TGF-β1 can increase expression of downstream cancer promotive factors, which forms molecular mechanisms of TGF-β1 contribution to tumourigenesis and progression of the malignant tumour.
In conclusion, this study reported that the miRNA-34b expression is down-regulated in cervical cancer samples and miRNA-34b can inhibit cell migration and induce cellular apoptosis through regulating TGF-β1. These results showed new viewpoints into the potential therapeutic effects of miRNA-34b in cervical cancer, demonstrating that miRNA-34b may be used as an effective biomarker as well as a treatment target for cervical cancer treatment in the future. The conclusions in this study should be verified by following in-vivo studies.
Disclosure statement
No potential conflict of interest was reported by the authors.
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Reference
- Silva GAF, Nunes RAL, Morale MG, et al. Oxidative stress: therapeutic approaches for cervical cancer treatment. Clinics (Sao Paulo) 2018;73:e548s.
- Gauri A, Messiah SE, Bouzoubaa LA, et al. Cervical cancer sociodemographic and diagnostic disparities in Florida: a population-based study (1981–2013) by stage at presentation. Ethn Health. 2018;1–9.
- Teoh D, Hultman G, DeKam M, et al. Excess cost of cervical cancer screening beyond recommended screening ages or after hysterectomy in a single institution. J Low Genit Tract Dis. 2018;22:184–188.
- Berman TA, Schiller JT. Human papillomavirus in cervical cancer and oropharyngeal cancer: one cause, two diseases. Cancer 2017;123:2219–2229.
- Fan Y, Meng Y, Yang S, et al. Screening of cervical cancer with self-collected cervical samples and next-generation sequencing. Dis Markers. 2018;2018:4826547.
- Mohr AM, Mott JL. Overview of microRNA biology. Semin Liver Dis. 2015;35:3–11.
- Rodrigues Lopes I, Silva RJ, Caramelo I, et al. Shedding light on microRNA function via microscopy-based screening. Methods. 2019;152:55–64.
- Ozbey U, Attar R, Romero MA, et al. Apigenin as an effective anticancer natural product: spotlight on TRAIL, WNT/beta-catenin, JAK-STAT pathways, and microRNAs. J Cell Biochem. 2018;120:1060–1067.
- Ren C, Liu Q, Wei Q, et al. Circulating miRNAs as potential biomarkers of age-related macular degeneration. Cell Physiol Biochem. 2017;41:1413–1423.
- Han MS, Lee JM, Kim SN, et al. Human papillomavirus 16 oncoproteins downregulate the expression of miR-148a-3p, miR-190a-5p, and miR-199b-5p in cervical cancer. Biomed Res Int. 2018;2018:1942867.
- Santos JMO, Peixoto da Silva S, Costa NR, et al. The role of microRNAs in the metastatic process of high-risk HPV-induced cancers. Cancers (Basel) 2018;10:493–508.
- Zhang L, Wang L, Dong D, et al. MiR-34b/c-5p and the neurokinin-1 receptor regulate breast cancer cell proliferation and apoptosis. Cell Prolif. 2019;52:e12527.
- Dou YD, Huang T, Wang Q, et al. Integrated microRNA and mRNA signatures in peripheral blood lymphocytes of familial epithelial ovarian cancer. Biochem Biophys Res Commun. 2018;496:191–198.
- Wang AM, Huang TT, Hsu KW, et al. Yin Yang 1 is a target of microRNA-34 family and contributes to gastric carcinogenesis. Oncotarget 2014;5:5002–5016.
- Hashemi M, Moazeni-Roodi A, Bahari G, et al. Association between miR-34b/c rs4938723 polymorphism and risk of cancer: an updated meta-analysis of 27 case-control studies. J Cell Biochem. 2019;120:3306–3314.
- Hashemi M, Hasanpour V, Danesh H, et al. Association between Pri-miR-34b/c rs4938723 polymorphism and bladder cancer risk. J Biomed Res. 2018;33:24–29.
- Revathidevi S, Manikandan M, Rao AK, et al. Analysis of APOBEC3A/3B germline deletion polymorphism in breast, cervical and oral cancers from South India and its impact on miRNA regulation. Tumour Biol. 2016;37:11983–11990.
- Cheung TH, Man KN, Yu MY, et al. Dysregulated microRNAs in the pathogenesis and progression of cervical neoplasm. Cell Cycle. 2012;11:2876–2884.
- Liu M, Wang Z, Liu Q, et al. Expression of micro-RNA-492 (MiR-492) in human cervical cancer cell lines is upregulated by transfection with wild-type P53, irradiation, and 5-fluorouracil treatment in vitro. Med Sci Monit. 2018;24:7750–7758.
- Dong J, Wang M, Ni D, et al. MicroRNA-217 functions as a tumour suppressor in cervical cancer cells through targeting Rho-associated protein kinase 1. Oncol Lett. 2018;16:5535–5542.
- Li H, Li X, Ge X, et al. MiR-34b-3 and miR-449a inhibit malignant progression of nasopharyngeal carcinoma by targeting lactate dehydrogenase A. Oncotarget 2016;7:54838–54851.
- Dong F, Lou D. MicroRNA-34b/c suppresses uveal melanoma cell proliferation and migration through multiple targets. Mol Vis. 2012;18:537–546.
- Xiao W, Zhou S, Xu H, et al. Nogo-B promotes the epithelial-mesenchymal transition in HeLa cervical cancer cells via r cells via Fibulin-5. Oncol Rep. 2013;29:109–116.
- Zhang F, Ren CC, Liu L, et al. HOXC6 gene silencing inhibits epithelial-mesenchymal transition and cell viability through the TGF-β/smad signaling pathway in cervical carcinoma cells. Cancer Cell Int. 2018;18:204.
- Wang A, Zhang Y, Cao P. Inhibition of BAP31 expression inhibits cervical cancer progression by suppressing metastasis and inducing intrinsic and extrinsic apoptosis. Biochem Biophys Res Commun. 2019;508:499–506.