Abstract
It has been found in several studies and research that the long non-coding RNA (Inc RNA) NEAT1 plays an important role in the development and succession in various malignant tumour. The development and metastasis of tumour mainly happen through lymph node. The purpose of this research is to explore the value of lymph node metastasis (LNM) in cancer. We have collected all the concerned studies about NEAT1 and researched the relationship between NEAT1 and lymph node metastasis. We have searched the studies by seeking database PubMed, Web of Science, and China National Knowledge Infrastructure (up to 10 January 2019), as well as a total of 821 patients from eight studies topics and made accordingly meta-analysis. By analyzing the data, we have found that the result is the high expression of NEAT1 associate with the metastasis of lymph node in different malignant tumours. The high level of NEAT1 expression can predict the metastasis of lymph node (OR = 3.36, 95% CI = 1.66–6.77, p = .000) and it is a molecular marker of positive lymph node for treatment.
Introduction
The incidence of malignant tumour is increasing year by year and the mortality of malignant tumour also appears uptrend. Although the medical level has been improved a lot than before, the survival rate is still low in many malignant tumours [Citation1]. So, it is rather urgent for us to find feasible biomarker for prevention, diagnosis therapies and especially the evaluation of lymph node metastasis and prognosis. Since some biomarker or sophisticated bioinformatics have appeared with non-matching staging system, and the ordinary staging system has already reached limited forecast accuracy, it is really urgent requested to develop more and more helpful forecasting instruments to satisfy various type of malignant tumour. The length of lnc RNA is longer than 200 nucleotide, which cannot be transferred into proteins [Citation2]. However, the Inc RNAs have been greatly used in the organismal pathway, and we could find out the actions, expressions and utterance of gene, including gene replacement, gene marking, gene alteration, selective splicing, cell cycle governance and passivation of primary tumour metastasis, all of which are important prognostic marker and metastasis sign [Citation3,Citation4].
The Nuclear paraspeckle assembly transcript 1 (NEAT1) gene has six transcripts [Citation5], and it is located at chromosome 11q13.1. The NEAT1 has been certified to be expressed in many kinds of tumor and has close relationship with the proliferation and invasion, like lung cancer, gastric cancer, breast cancer, oesophagus cancer and endometrial cancer [Citation6–10]. Therefore, we have collected lots of articles and studies for research, and the relationship between lymph node metastasis and NEAT1 will direct the measurable meta-analysis.
Materials and method
Publication search
We took careful research with the data about biomarker for lymph node metastasis in PubMed, Web of Science and China National Knowledge Infrastructure (CNKI) database (till up to 10 January 2019). And, our research list was (“NEAT1 OR Nuclear paraspeckle assembly transcript 1”) AND (“cancer OR tumour OR neoplasm”). The relevant references of concerned article were researched to acquire other latent studies, and all of studies were collected by our meta-analysis to probe the relationship between NEAT1 and lymph node metastasis ().
Inclusion and exclusion criteria
We have made the Inclusion criteria, and they were as following: (1) the expression criterion of lnc RNA NEAT1 in tumour tissue can be estimated. (2) Articles probing as the part of NEAT1 in the exploitation of tumour which can be different biological characteristics. (3) Patients were allocated as high expression and low expression based upon the expression criteria of NEAT1, the criteria is the expression of NEAT1 was detected using qRT-PCR and standardized to GAPDH or β-actin between absent and present of lymph node metastasis. (4) All of diagnosis of LNM was depend on pathology. (5) Important data were contained for the evaluation of Odds Ratios (OR) and corresponding 95% CI. And, the designing exclusion criteria were as follows: (1) articles researching the molecular configuration and functions of NEAT1. (2) The manner of metastasis was only limited by lymphonode except for other metastasis manner. (3) Editorials, letters, expert opinions, reviews and case reports; (4) studies without practicable data(5) reproductive publications.
Data extraction
The two researchers (ZZ, LZ) extracted the data from effective studies, following with the same inclusion and exclusion criteria, the disagreement parts were solved by other authors (ZZ, LZY). The information was concentrated from these studies: (1) first author, country, data of publication, tumour type, total number of patients, different LNM group of high NEAT1 expression and low NEAT1 expression. (2) Total number patients with LNM in each group. (3) The detection approach of NEAT1 expression level and cut-off value.
Statistical method
We analyzed the data using Stata SE12.0 (Stata Corporation) and conducted the test of the heterogeneity of combined ORs using Cochran’s Q test and Higgins I-squared statistic. During Q test, we considered it is important for our studies when p values of heterogeneity <.05 and I2 values >50%. If the heterogeneity was important, we would use the random effects model. We evaluated the latent publication bias though the Egger’s test, and the stability of the results through sensitivity analysis.
Result
Research characteristic
All of eleven studies were selected effectively under applicable criteria. Finally, we have involved eight studies under accurate analysis [Citation7–9,Citation11–15], with 821 patients involved from China in the studies. There were seven types of cancer in these studies, including one colorectal cancer, one oesophagus cancer, two lung cancer, one breast cancer, one endometrial cancer, one pancreatic cancer and one renal cancer. All of characteristics in this study are outlined in . The diagnosis of patients was mainly based on the pathology, and no one accepted any radiotherapy or chemotherapy before operation. These studies can be divided into high expression group and low expression group by following designed method: (1) the high NEAT1 group with NEAT1 expression levels > median value and the low NEAT1 group with NEAT1 expression levels < median value. (2) The level of dividing into group is based on the NEAT1 expression and cut off value.
Table 1. Summary of the eight included studies.
Result of meta-analysis
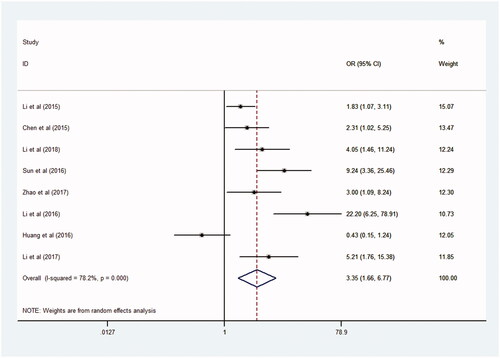
The result of this meta-analysis with high NEAT1 expression was associated with high incidence of lymph node metastasis, and an important heterogeneity was certified from the studies (I2 = 78.2%, p = .000), and then the random-effects model was adopted. And, old ratios for high NEAT1 expression versus low NEAT1 expression was 3.36 (95% CI = 1.66–6.77, p = .000, ). By comparing with the effect of lymph node metastasis between high NEAT1 expression group and low HEAT1 expression group, we found that higher NEAT1 expression is fatidic with higher LNM (). And, the result showed that malignant tumour patients with high NEAT1 expression in tumour tissues were much more prone to become LNM.
Publication bias
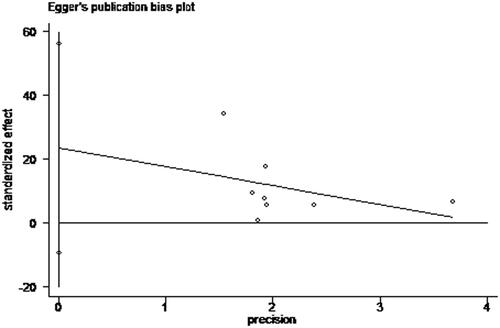
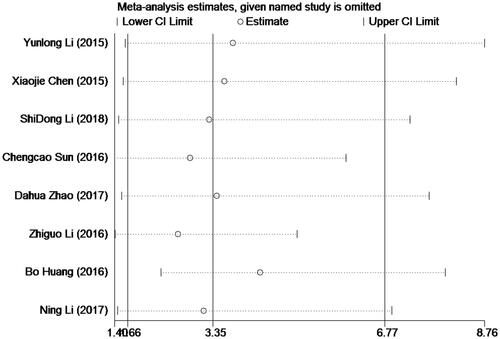
The publication bias of the current meta-analysis was valued by Egger’s test (). And, no more important publication bias was surveyed for it. We took statistically analysis to the bias of this meta-analysis through the Egger's test, and the results of Egger's test (p = .130) indicated with no publication bias (p > .05). We detected the heterogeneity through sensitivity analysis, and we can conduct the result is stable without deleting any study ().
Discussion
Lnc RNA for ENCODE project has drawn extensive attention. IncRNA is non-coding RNA with more than 200 nt longer in length, which was first found out by Brockdorff et al. [Citation16]. There has been plenty of research concerning about lnc RNA on the progression of diagnosis and treatment about malignant cancer in recent years [Citation17]. On the other side, more and more evidence have indicated that cancer-specific lncRNA might serve as potent prognostic molecular target [Citation18,Citation19]. So, the general identification of lncRNA associating with cancer may help us reach the path breaking manners, and make exactly analyze and prediction about the prognosis of various of cancer.
The first prognostic marker of lung cancer metastasis was the lncRNA MALAT1, which is associated with other human tumour metastasis [Citation20]. The nuclear paraspeckle assembly transcript 1 (NEAT1) is a newly identified nuclear-restricted long non-coding RNA. In recent years, the function and action of lncRNA NEAT1 have been found in different types of cancers [Citation7–9,Citation11–15], and the general mechanism in the future between malignant tumour, and NEAT1 is still developing continuously. As an architectural lncRNA in nuclear paraspeckles, lncRNA NEAT1 is necessary for paraspeckle completeness and functions [Citation21]. It was found by Adriaens et al. that DNA damage agent could cause NEAT1 expression and paraspeckle formation through manner of p53-dependent, conducing to the chemo-resistance in malignant tumour cell [Citation22]. Furthermore, NEAT1 has devised isolate paraspeckle proteins into the nuclear bodies, restricting the transcriptional activation or inactivation activity of these proteins in the nucleoplasm [Citation23]. Li et al. have found lncRNA NEAT1 promotes proliferation and invasion via targeting miR-181a-5p in non-small cell lung cancer [Citation13]. Zhao et al. showed that NEAT1 negatively regulated miR-218 expression and has promoted breast cancer’s progression [Citation7]. Huang has revealed lncRNA NEAT1 facilitates in pancreatic cancer’s progression through negative modulation of miR-506-3p [Citation14]. Su et al. found NEAT1 was a kind of prognosis biomarker and it regulated cancer’s progression via epithelial-mesenchymal transition in clear cell renal of cell carcinoma [Citation15]. Based on the research of Tao Chen et al. and Yunyuan Zhang et al. [Citation24,Citation25], we have found lncRNA NEAT1 was an important prognosis biomarker, which indicates that the high NEAT1 expression showed important association with the poor OS, however, he only explored the relationship between lncRNA NEAT1 and LNM by four research papers and they could not confirm any association between NEAT1 expression and LNM (pooled OR: 2.28, 95% CI: 0.72–7.26, p = .161). The conclusion is not persuasive obviously; as a result, we took further research and studies for the recent four years to prove the association between lncRNA NEAT1 and LNM. The analytical reports have shown a strong relationship between NEAT1 and LNM (OR = 3.36, 95% CI = 1.66–6.77, p = .000). The clear realization of the relation between NEAT1 and LNM help us know more about cancer. In conclusion, lncRNA NEAT1 can be a significant biomarker for predicting and evaluating the possibility of lymph node metastasis for patients’ treatment, and what is more, it is a useful technique and signal for clinical treatment and achieving effective therapy.
Lymphatics may serve as starting point of access to the systemic circulation of tumour cells. Malignancies of ectodermal or endodermal origin may spontaneously migrate to lymph nodes under favourable tumour microenvironment, and the underlying mechanisms have been a mooting point in scientific enquiry of cancer researches. Although the precise mechanisms remained unrevealed, clinical and basic science studies suggest that molecular receptors unique to lymphatic endothelial cells, such as chemokines, Prox1, VEGF receptors, integrin, podoplanin, angiopoietin-1 and CCL21 [Citation26–30]. Therefore, the association between NEAT1 and lymph node metastasis is one of the many façades of molecular pathological mechanisms. Another limitation of this meta-analysis resides in the small number of literature available for aggregated analysis so that the heterogeneity between studies cannot be assessed adequately. Finally, the definition of high NEAT1 and low NEAT1 expression differs in each study, compromising the reliability of the conclusion of this study.
Author contributions
Author Zhenglun Yu and Lei Zhang wrote the main manuscript text, and author Lei Zhangand Zhe Zhang prepared , and author Zhenglun Yu prepared . All authors reviewed the manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30.
- Knauss JL, Sun T. Regulatory mechanisms of long noncoding RNAs in vertebrate central nervous system development and function. Neuroscience. 2013;235:200–214.
- Loewen G, Jayawickramarajah J, Zhuo Y, et al. Functions of lncRNA HOTAIR in lung cancer. J Hematol Oncol. 2014;7:90.
- Iguchi T, Uchi R, Nambara S, et al. A long noncoding RNA, lncRNA-ATB, is involved in the progression and prognosis of colorectal cancer. Anticancer Res. 2015;35:1385–1388.
- Aken BL, Ayling S, Barrell D, et al. The Ensembl gene annotation system. Database (Oxford). 2016;2016:1–19.
- Fu JW, Kong Y, Sun X. Long noncoding RNA NEAT1 is an unfavorable prognostic factor and regulates migration and invasion in gastric cancer. J Cancer Res Clin Oncol. 2016;142:1571–1579.
- Zhao D, Zhang Y, Wang N, et al. NEAT1 negatively regulates miR-218 expression and promotes breast cancer progression. Cbm. 2017;20:247–254.
- Chen X, Kong J, Ma Z, et al. Up regulation of the long non-coding RNA NEAT1 promotes esophageal squamous cell carcinoma cell progression and correlates with poor prognosis. Am J Cancer Res. 2015;5:2808–2815.
- Li Z, Wei D, Yang C, et al. Overexpression of long noncoding RNA, and NEAT1 promotes cell proliferation, invasion and migration in endometrial endometrioid adenocarcinoma. Biomed Pharmacother. 2016;84:244–251.
- Lu Y, Li T, Wei G, et al. The long non-coding RNA NEAT1 regulates epithelial to mesenchymal transition and radioresistance in through miR-204/ZEB1 axis in nasopharyngeal carcinoma. Tumor Biol. 2016;37:11733–11741.
- Li YL, Li YH, Chen WP, et al. NEAT expression is associated with tumor recurrence and unfavorable prognosis in colorectal cancer. Oncotarget. 2015;6:27641–27650.
- Sun C, Li S, Zhang F, et al. Long non-coding RNA NEAT1 promotes non-small cell lung cancer progression through regulation of miR-377-3p-E2F3 pathway. Oncotarget. 2016;7:51784–51814.
- Li SD, Yang JM, Xia YB, et al. Long noncoding RNA NEAT1 promotes proliferation and invasion via targeting miR-181a-5p in non-small cell lung cancer. Oncol Res. 2018;26:289–296.
- Huang B, Liu C, Wu Q, et al. Long non-coding RNA NEAT1 facilitates pancreatic cancer progression through negative modulation of miR-506-3p. Biochem Biophys Res Commun. 2017;482:828–834.
- Li N, Li ZG, Wei DJ, et al. LncRNA, NEAT1 is a prognosis biomarker and regulates cancer progression via epithelial-mesenchymal transition in clear cell renal cell carcinoma. Cancer Biomark. 2017;19:75–83.
- Brockdorff N, Ashworth A, Kay GF, et al. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell. 1992;71:515–526.
- Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–361.
- Hong HH, Hou LK, Pan X, et al. Long non-coding RNA UCA1 is a predictive biomarker of cancer. Oncotarget. 2016;7:44442–44447.
- Cui Z, Chen Y, Xiao Z, et al. Long noncoding RNAs as auxiliary biomarkers for gastric cancer screening: a pooled analysis of individual studies. Oncotarget. 2016;7:25791–25800.
- Gutschner T, Hammerle M, Diederichs S. MALAT1 – a paradigm for long noncoding RNA function in cancer. J Mol Med. 2013;91:791–801.
- Bond CS, Fox AH. Paraspeckles: nuclear bodies built on long noncoding RNA. J Cell Biol. 2009;186:637–644.
- Adriaens C, Standaert L, Barra J, et al. p53 induces formation of NEAT1 lncRNA-containing paraspeckles that modulate replication stress response and chemosensitivity. Nat Med. 2016;22:861–868.
- West JA, Mito M, Kurosaka S, et al. Structural, super-resolution microscopy analysis of paraspeckle nuclear body organization. J Cell Biol. 2016;214:817–830.
- Chen T, Yang H, Wang P, et al. Prognostic role of long noncoding RNA NEAT1 in various carcinomas: a meta-analysis. Onco Targets Ther. 2017;10:993–1000.
- Zhang YY, Lun LM, Li H, et al. The value of lncRNA NEAT1 as a prognostic factor for survival of cancer outcome: a meta-analysis. Sci Rep. 2017;10:1–9.
- Oliveiraneto HH, Souza PPC, Da Silva MRB, et al. The expression of chemokines CCL19, CCL21 and their receptor CCR7 in oral squamous cell carcinoma and its relevance to cervical lymph node metastasis. Tumor Biol. 2013;34:65–70.
- Ramani P, Norton A, Somerville M, et al. PROX1 lymphatic density correlates with adverse clinicopathological factors, lymph node metastases and survival in neuroblastomas. J Neurooncol. 2012;108:375–383.
- Guo J, Lou W, Ji Y, et al. Effect of CCR7, CXCR4 and VEGF-C on the lymph node metastasis of human pancreatic ductal adenocarcinoma. Oncol Lett. 2013;5:1572–1578.
- Ganguly KK, Pal S, Moulik S, et al. Integrins and metastasis. Cell Adh Migr. 2013;7:251–261.
- Huber GF, Fritzsche FR, Züllig L, et al. Podoplanin expression correlates with sentinel lymph node metastasis in early squamous cell carcinomas of the oral cavity and oropharynx. Int J Cancer. 2011;129:1404–1409.