Abstract
Objective
The current study aimed to explore the function of miR-638 on the progression of oral squamous cell carcinoma (OSCC) and relevant molecular mechanisms.
Methods
Expression profile of miR-638 in OSCC tissues and cells was detected using quantitative real-time polymerase chain reaction (qRT-PCR) method. Chi-square test was performed to estimate the relationship between miR-638 and clinical parameters of OSCC cases. Cell viability and motility abilities were estimated using MTT and transwell assays, respectively. Potential targets of miR-638 in OSCC were identified through bioinformatics analysis and luciferase reporter assay.
Results
MiR-638 exhibited decreased expression in OSCC tissues and cells, compared to non-cancerous controls (P < .05 for both). Moreover, its down-regulation was closely correlated with lymph node metastasis (P = .044) and TNM stages (P = .001). Enforced miR-638 expression reduced cell proliferation, migration and invasion, while its knockdown exhibited opposite effects. Phospholipase D1 (PLD1) was confirmed as a target of miR-638 in OSCC. MiR-638 could inhibit wnt/β-catenin pathway through targeting PLD1, thus realizing its anti-tumour action in OSCC.
Conclusion
MiR-638 may be a tumour suppressor in OSCC by targeting PLD1/Wnt/β-catenin pathway.
Introduction
Oral squamous cell carcinoma (OSCC) is a frequently diagnosed malignant head and neck cancer, especially among men [Citation1,Citation2]. Tobacco and alcohol are two major risk factors for OSCC [Citation3]. Currently, surgery is the primary therapeutic strategy for OSCC, followed by radiotherapy and chemotherapy [Citation4]. Treatments have significantly improved in recent years, but the prognosis of OSCC cases is still unsatisfactory, especially among those in advanced stages, showing a five-year survival rate of less than 50% [Citation5]. Delay in diagnosis, high invasion, and soaring costs for therapy may be partially responsible for dismal clinical outcomes of OSCC [Citation6,Citation7]. And ascertaining molecular etiology of OSCC is of great help for early diagnosis and treatment of this disease. Reportedly, OSCC is a combined result of multiple genetic alterations and environmental factors [Citation8]. So genetic factors play a key role in the pathogenesis of OSCC.
MicroRNAs (MiRNAs) are a group of single-strand non-coding RNAs, with a length of about 22 nucleotides [Citation9]. Until now, ∼2000 miRNAs have been identified in humans, which could regulate one-third of genes in the genome [Citation10]. Given their regulatory functions on gene expression, miRNAs impose certain influences on a variety of biological pathways [Citation11]. Expression patterns of miRNAs are specific towards different cells, tissues and developmental stages [Citation12], and their abnormal expressions may cause gene dysregulation, such as translation repression, direct mRNA degradation, and miRNA-mediated mRNA decay, thus resulting in diseases [Citation13]. Dysregulated miRNAs may act as oncogenes or suppressors in tumourigenesis. MiR-638 is a widely studied miRNA, and its down-regulation has been observed in several types of malignancies, such as hepatocellular carcinoma [Citation14], osteosarcoma [Citation15], gastric cancer [Citation16], etc. MiR-638 expression deficiency may contribute to malignant progression. However, the functional roles of miR-638 in OSCC were rarely reported.
In this study, we investigated expression patterns of miR-638 in OSCC tissues and cells. Moreover, cell experiments were carried out to explore the function of miR-638 on OSCC progression, as well as related molecular mechanisms.
Materials and methods
Patients and tissue specimens
A total of 107 OSCC patients who were pathologically confirmed were recruited from the School and Hospital of Stomatology, Shandong University. Malignant tissue specimens and adjacent normal ones were collected from all the cases in surgery, frozen in liquid nitrogen immediately, and then stored at −80 °C. All of the included patients were adults, and none of them had received any pre-operative treatments, such as chemotherapy and radiotherapy.
Clinical characteristics of the included patients were collected and summarized in . The cases contained 60 males and 47 females, with an average age of 62.08 ± 10.25 years. Of them, 54 had a history of smoking, while 55 had a drinking history. Besides, 45 patients exhibited low differentiation, and lymph node metastasis was observed in 38 ones. According to TNM staging, 65 patients were classified into stages I–II while other 42 into stages III–IV.
Table 1. The Association of miR-638 expression with clinical characteristics of OSCC patients.
The present study was approved by the Ethics Committee of the hospital. All of the patients signed written informed consents.
Cell line and culture
OSCC cell lines Tca-8113, SCC-4 and SCC-9 (ATCC® CRL-1629™) and normal human immortalized oral mucosa epithelial cell HOK (human oral keratinocytes, NHOK) (ATCC® PCS-200–014™) were bought from American type culture collection (ATCC). All of the cell lines were cultured in DMEM medium (Gibco, USA) supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientifc, Inc., Waltham, MA, USA). The cell medium was incubated at 37 °C with the presence of 5% CO2.
RNA isolation and quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was extracted from the collected tissue and cell specimens using Trizol reagent (Invitrogen, Thermo Fisher Scientific, Inc.), and experimental procedures were carried out according to the introduction of the manufacturer. Then cDNA was synthesized using PrimerScript RT reagent kit (Takara, Dalian, China). Quantitative analysis was carried out using the qRT-PCR method adopting SYBR Green PCR master mix (Applied Biosystems, USA) in 7300 Real-Time PCR System (Applied Biosystems, USA). Our used primer sequences were as follows: U6 forward: 5′-CTCGCTTCGGCAGCACA-3′; reverse: 5′-AACGCTTCACGAATTTGCGT-3′, miR-638 forward: 5′-AAGGGATCGCGGGCGGGT-3′; reverse: 5′-CAGTGCAGGGTCCGAGGT-3′; GAPDH forward: 5′-AAGGCTGGGGCTCATTTGCAGG-3′; reverse: 5′-AGTTGGTGGTGCAGGAGGCA-3′, phospholipase D1 (PLD1) forward: 5′-AAGGCGGCTCGTGATGTGG-3′; reverse: 5′-ATGGGCTGTTGTTTGAGACTTTGG-3′. U6 was employed as an internal control for miRNA detection, while GAPDH for mRNA detection. Expression levels of the genes were calculated using 2−ΔΔCt formula. Each test was repeated at least three times.
Cell transfection
To investigate the roles of miR-638 in OSCC progression and related molecular mechanisms, miR-638 mimic, miR-638 inhibitor, si-PLD1 and corresponding negative controls were designed and synthesized by HANBIO company (Shanghai, China). For transfection, the cells were harvested at the logarithmic phase and digested using 0.25% trypsin. Then, the cells were seeded into a 6-well plate in a density of 1 × 105 cells/mL. Later, Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.) was used for cell transfection. Subsequently, the transfected cells were incubated at 37 °C with 5% CO2 for 48 h. Relative expressions of the genes were detected using qRT-PCR method to estimate transfection efficacy.
Western blot analysis
Western blot analysis was performed to detect expression patterns of related proteins. RIPA Lysis and Extraction Buffer (Thermo Scientific, Waltham, MA, USA) were used for protein isolation. Then, the protein was quantified adopting BCA Protein Assay Kit (Thermo Scientific, Waltham, MA, USA). Same volumes of protein samples were loaded to 10% SDS-PAGE, and then transfected to polyvinylidene fluoride membrane (0.45 µm pore size; EMD Millipore, Billerica, MA, USA) under the condition of 300 mA for 1.5 h. Later, 5% skimmed milk was used to block the membrane at room temperature for 2 h. Subsequently, the membranes were incubated with specific antibodies, respectively, such as anti-β-catenin antibody (1:5000, Abcam), anti-C-myc antibody (1:1000, Abcam), anti-cyclin D1-antibody (1:10,000, Abcam), and anti-GAPDH-antibody (1:10,000, Abcam). GAPDH was employed as a loading control. Then, the membranes were incubated with secondary anti-rabbit IgG antibody (1:2000, Abcam) at room temperature for 2 h. Chemi Genius gel imaging system was used to analyze band gray. Each experiment was repeated three times.
Cell proliferation evaluation
The effects of miR-638 expression levels on cell proliferation were investigated using MTT method. The cells were seeded into 96-well plate with a destiny of 1 × 104 cells/mL, and then incubated at 37 °C with 5% CO2. Later, 20 μL 3–(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) (Sigma) was added into the cell medium every 24 h (cultured for 0, 24, 48 and 72 h), and incubated for additional 4 h. Afterwards, 150 DMSO was added and incubated at dark for 30 min to stop the reaction. The absorbance at 490 nm of the cell medium was detected using a Microplate Reader (TECAN, Salzburg, Austria) to estimate cell proliferation ability. Each test was performed three times.
Cell motility assay
Transwell analysis (8.0 µm pore size, Costar, Shanghai, China) was performed to estimate cell motility abilities, including migration and invasion. For migration analysis, the upper chamber was filled with 200 μL serum-free DMEM medium, while the lower chamber was coated with 500 μL DMEM medium with 10% FBS. The cells (5 × 104 cells/mL) were seeded into the upper chamber, and incubated 37 °C with 5% CO2. 48 h later, the cells in the lower chamber were counted under an inverted microscope (IX31; Olympus Corporation, Tokyo, Japan). Five random files were selected for each sample. In invasion assay, the upper chamber was pre-coated with additional Matrigel (Corning Glass Works, Corning, N.Y., USA).
Luciferase reporter assay
Luciferase reporter assay was performed to explore the targeted relationship between miR-638 and PLD1. In brief, the fragment of PLD1 gene containing the binding site of miR-638 (PLD1-wt), and the fragment with the mutated binding site (PLD1-mt) were amplified through PCR method and cloned to luciferase reporter vector pGL3 (Promega, USA) respectively. Experiment procedures were carried out according to the manufacturer’s instruction. The recombinational luciferase vectors (PLD1-wt and PLD1-mt) combined with miR-638 mimic or mimic were transfected to OSCC cells in vitro using Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.). After incubation for 48 h, luciferase activity of the transfected cells was analyzed employing Dual-Luciferase Reporter Assay System (Promega Corporation). Each sample was tested in triplicate.
Statistical analysis
Continuous variables were shown as mean ± standard deviation (SD), and compared between two groups using student’s t-test. Categorical data were recorded as case number and percentage, and their comparisons were performed through the chi-square test. All data analyses were performed using SPSS 18.0 software (SPSS, Inc., Chicago, IL, USA), and figures were plotted applying GraphPad Prism version 5.0 (GraphPad, San Diego, CA, USA). P-values <.05 predicted that the results were statistically significant.
Results
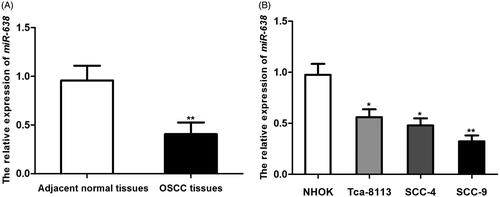
Down-regulation of miR-638 in OSCC tissues and cells
The expression of miR-638 in collected OSCC tissues and cells was detected using qRT-PCR method. As shown in , the expression level of miR-638 mRNA in OSCC tissues was significantly decreased compared with corresponding adjacent normal tissues (P < .001, ). Moreover, miR-638 expression level in OSCC cells Tca-8113, SCC-4 and SCC-9 were all significantly lower than that in HNOK cells (, P < .05 for all), with lowest figure in SCC-9 (P < .001).
Association of miR-638 expression with clinical characteristics of OSCC patients
The mean values of miR-638 expression in malignant specimens were frequently employed as the cut-off value for grouping in the analysis of the relationship between miR-638 expression and clinical characteristics among cancer patients in previous studies [Citation17,Citation18]. In our study, the patients were divided into high (n = 59) and low (n = 48) expression group based on their mean expression of miR-638 in HCC tissues. Chi-square test was applied to estimate the relationship between miR-638 expression and clinical characteristics of OSCC patients. The results demonstrated that miR-638 expression was negatively correlated with lymph node metastasis (P = .044) and TNM stages (P = .001). Meanwhile, the patterns of miR-638 expression had no close relationship with the patients’ age (P = .131), gender (P = .414), smoking (P = .281), drinking (P = .899), or differentiation (P = .133) ().
MiR-638 inhibited OSCC cell proliferation, migration and invasion
In order to investigate functional roles of miR-638 in OSCC progression, miR-638 mimic, miR-638 inhibitor, as well as corresponding negative controls were designed and transfected to OSCC cells in vitro. QRT-PCR method was performed to estimate transfection efficacy. Analysis results demonstrated that compared to the controls, the transfection of miR-638 mimic could significantly enhance the expression of miR-638 (P < .01), while miR-638 inhibitor could suppress miR-638 expression in OSCC cells (P < .05) ().
Figure 2. The transfection with miR-638 mimic could enhance the expression of miR-638, while transfection with miR-638 inhibitor down-regulated miR-638 (A). Enforced miR-638 could significantly suppress OSCC cells’ proliferation (B), migration (C) and invasion (D), and the knockdown of miR-638 promoted malignant behaviors of OSCC cells. **P < .01; *P < .05.
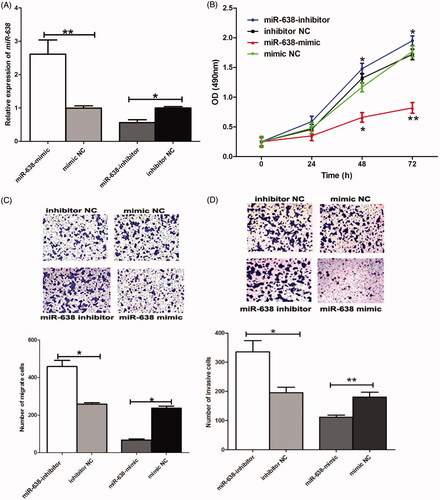
Biological behaviors of the transfected cells were detected. MTT assay suggested that enforced expression of miR-638 using mimic transfection could significantly suppress cell proliferation (P < .05); moreover, the knockdown of miR-638 enhanced proliferation ability of OSCC cells (P < .05) (). Furthermore, restored expression of miR-638 imposed inhibiting effects on cell migration and invasion, while the transfection of miR-638 inhibitor could promote cell migration and invasion, according to transwell analysis (P < .05 for all) ().
PLD1 was a potential target of miR-638 in OSCC
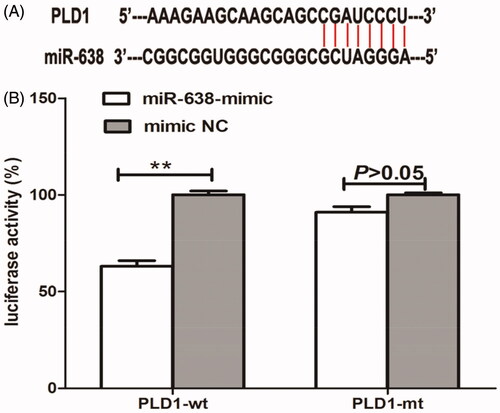
Bioinformatics analysis implemented through TargetScan Human prediction software demonstrated that the 3′UTR of PLD1 gene had complementary sequences of miR-638 (). Moreover, the study carried out in gastric cancer also revealed a targeted relationship between PLD1 and miR-638 [Citation16]. Thus, we speculated that PLD1 was a potential target of miR-638 in OSCC. Luciferase reporter assay demonstrated that in cells transfected by PLD1-wt, the presence of miR-638 mimic could significantly reduce luciferase activity of the cells (P < .01), while the co-transfection with miR-638 and PLD1-mt had no obvious influences on luciferase activity of the cells (P > .05) (). MiR-638 could bind to the 3’ UTR of PLD1 gene in OSCC.
Figure 3. Bioinformatics analysis demonstrated that the 3′UTR of PLD1 gene had complementary sequences of miR-638 (A). In cells transfected by PLD1-wt, the presence of miR-638 mimic could significantly reduce luciferase activity of the cells, while the co-transfection with miR-638 and PLD1-mt had no obvious influences on luciferase activity of the cells (B). **P < .01.
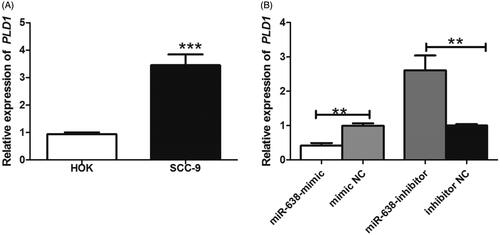
QRT-PCR method was used to investigate the relative expression of PLD1 mRNA in OSCC. The results suggested that the levels of PLD1 mRNA were significantly increased in OSCC cells, compared to non-cancerous cells (P < .001) (). Furthermore, enforced expression of miR-638 could obviously suppress PLD1 expression (P < .01), while miR-638 inhibition up-regulated PLD1 in OSCC (P < .01) (). MiR-638 could negatively regulate the expression of PLD1 in OSCC. PLD1 was a target of miR-638 in OSCC.
MiR-638 could regulate the activity of wnt/β-catenin signalling pathway
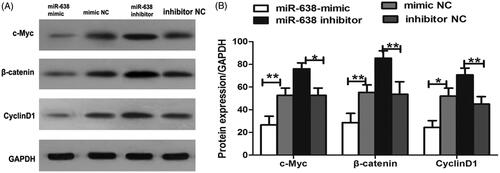
Western blot analysis was performed to investigate the expression of the proteins in wnt/β-catenin signalling pathway for the transfected cells. As displayed in , compared to the controls, the levels of c-Myc, β-catenin and Cycline D1 were significantly decreased after the transfection of miR-638 mimic, while the transfection of miR-638 inhibitor could obviously constrain the expression of the proteins (P < .05 for all).
Molecular mechanisms of miR-638 in the progression of OSCC
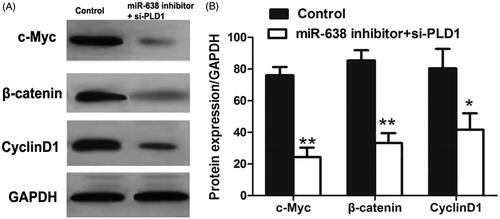
In order to investigate potential mechanisms underlying functional roles of miR-638 in OSCC progression, we designed si-PLD1 vector to knockdown PLD1 in cells transfected by miR-638 inhibitor, and cells transfected by miR-638 inhibitor served as controls. Western blot analysis suggested that compared to the controls, the co-transfection with miR-638 inhibitor and si-PLD1 could down-regulate c-Myc, β-catenin and Cycline D1, revealing the inactivity of wnt/β-catenin signalling pathway (P < .05 for all) ().
Figure 6. Western blot analysis for proteins in wnt/β-catenin pathway after the co-transfection with miR-638 inhibitor and si-PLD1 (A). Quantitative analysis for western blot results (B). **P < .01; *P < .05.
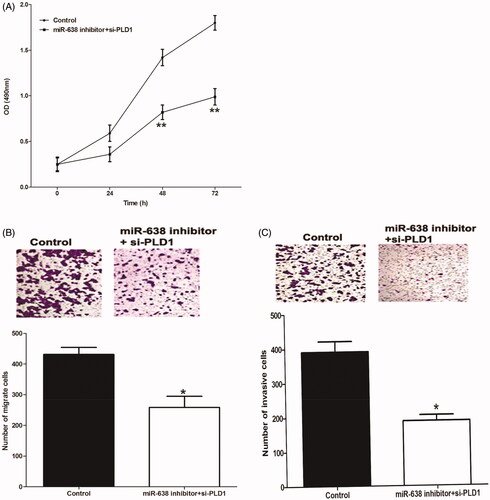
Biological behaviors of OSCC cells transfected by miR-638 inhibitor and si-PLD1 were also investigated in the current study. MTT assay suggested that the co-transfection with miR-638 inhibitor and si-PLD1 decreased cell proliferation ability (P < .01) (). Transwell analyses demonstrated that the presence of si-PLD1 could obviously suppress the migration () and invasion () of OSCC cells transfected by miR-638 inhibitor (P < .05 for both).
Discussion
MiRNAs play important roles in tumourigenesis through their target genes. Exploring their function and related molecular mechanisms may provide new insight into the etiology of cancers. In the current study, we analyzed the functional roles of miR-638 in OSCC progression, as well as, potentially related molecular mechanisms. We found that the expression of miR-638 was significantly decreased in OSCC tissues and cells; moreover, its down-regulation predicted positive lymph node metastasis and advanced TNM stages. The expression of miR-638 was negatively correlated with malignant behaviors of OSCC cells in vitro. MiR-638 could suppress the wnt/β-catenin signalling pathway through PLD1, thus inhibiting OSCC progression.
OSCC is a frequently diagnosed malignancy around the world, with poor five-year survival [Citation19]. And exploring molecular mechanisms underlying OSCC progression is crucial to improve its treatments and prognosis. MicroRNAs, gene expression regulators, are involved in various types of cancers, including OSCC [Citation20]. Dysregulated miRNAs are reportedly to act as tumour suppressors or oncogenes in OSCC origination and progression. In the current study, we found that the expression of miR-638 was significantly down-regulated in OSCC tissues and cells; moreover, its decreased expression predicted positive lymph node metastasis and advanced TNM stages. MiR-638 expression showed a negative correlation with OSCC aggressive progression. Furthermore, cell experiments demonstrated that enforced expression of miR-638 could inhibit malignant proliferation, migration and invasion of OSCC cells, while its knockdown might enhance aggressive biological behaviors of OSCC cells. All of the data revealed that miR-638 acted as a tumour suppressor in OSCC. The present study might be the first one investigating the functional roles of miR-368 in OSCC. Our findings were in consistent with those from studies on other types of cancers. Shen et al. reported that miR-638 as a tumour suppressor played pivotal roles in gastric cancer [Citation21]. Shi et al. suggested that hepatocellular carcinoma patients with low expression of serum exosomal miR-638 were more likely to undergo poor prognosis. Moreover, the proliferation ability of hepatocellular carcinoma cells was inhibited after the transfection with miR-638 mimic [Citation22]. Taken together, miR-638 played an inhibitory role in tumourigenesis and thus might be employed as a therapeutic target.
MiRNAs possess no capability to code protein, and they take part in biological processes through their target genes [Citation23]. In the current study, PLD1 was confirmed as a target gene of miR-638, according to bioinformatics analysis and luciferase reporter assay. PLD1 expression was negatively regulated by miR-638. Such conclusion was in line with that from the study of Zhang et al [Citation16]. In their study, PLD1 was confirmed as a target of miR-638 as well, and miR-638 inhibited the progression of gastric cancer through regulating PLD1 expression. PLD1 belongs to PLD lipid-signalling enzyme super-family, which represents a major source of signal-activated phosphatidic acid (PtdOH) generation for numerous cell-surface receptors [Citation24]. PLD1 is generally accepted as an oncogene, and its up-regulation promotes angiogenesis and metastasis in tumourigenesis [Citation25]. Oncogenic function of PLD1 has been reported in multiple malignancies, including glioma [Citation26], hepatocellular carcinoma [Citation27], breast cancer [Citation28], etc. In the current study, we confirmed that miR-638 played an anti-tumour role in OSCC through its negative regulation on PLD1.
Several published articles have demonstrated that PLD1 could activate wnt/β-catenin in cancer progression [Citation29–31]. Based on these articles, we speculated that miR-638 might regulate the wnt/β-catenin pathway through PLD1. In our study, we found that enforced expression of miR-638 could inactivate wnt/β-catenin, while miR-638 knockdown resulted in excessive activation of the pathway. PLD1 can reverse the function of miR-638 on wnt/β-catenin pathway, and inhibitory function of miR-638 on wnt/β-catenin pathway is dependent on PLD1. The present study might be the first one discussing the functional roles of miR-638 in OSCC progression and related molecular mechanisms. Despite those encouraging results, several limitations in this work should be stated. Firstly, the sample size was relatively small, which might influence the statistical power of our results. Secondly, the function of miR-638 was proved only in one OSCC cell line due to the limited study period. Expression profiles and function of miR-638 in other OSCC cell lines remained unclear. SCC-9 is a tongue squamous cell carcinoma cell line which was derived from a 25-year-old male, and shows high colony formation ability in vitro and oncogenicity in vivo [Citation32]. This cell line is frequently used for OSCC experiments in vitro. Thus, the results obtained in our study had certain representativeness. Thirdly, animal models are required to verify our results. Besides, apart from wnt/β-catenin pathway, miR-638 might take part in OSCC progression through multiple signalling pathways. Therefore, further investigations should be designed to explore regulation through netweb for miR-638 in OSCC.
In conclusion, the expression of miR-638 is down-regulated in OSCC, and shows a negative correlation with cancer progression. MiR-638 can inhibit the proliferation, migration, and invasion of OSCC cells. The anti-tumour action of miR-638 in OSCC may be dependent on miR-638/wnt/β-catenin axis.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA: A Cancer J Clinic. 2017;67:7–30.
- Rivera C. Essentials of oral cancer. Int J Clin Exp Pathol. 2015;8:11884–11894.
- Kumar M, Nanavati R, Modi TG, et al. Oral cancer: etiology and risk factors: a review. J Cancer Res Ther. 2016;12:458–463.
- Ettinger KS, Ganry L, Fernandes RP. Oral cavity cancer. Oral and Maxillofacial Surg Clin N Am. 2019;31:13–29.
- Huang SH, O'Sullivan B. Oral cancer: current role of radiotherapy and chemotherapy. Med Oral Patol Oral Cir Bucal. 2013;18:e233–40.
- Speight PM, Farthing PM. The pathology of oral cancer. Br Dent J. 2018;225:841–847.
- Huber MA, Tantiwongkosi B. Oral and oropharyngeal cancer. Med Clin North Am. 2014;98:1299–1321.
- Irimie AI, Ciocan C, Gulei D, et al. Current insights into oral cancer epigenetics. IJMS. 2018;19:670.
- Li Z, Xu R, Li N. MicroRNAs from plants to animals, do they define a new messenger for communication? Nutr Metab (Lond). 2018;15:68.
- Hammond SM. An overview of microRNAs. Adv Drug Deliv Rev. 2015;87:3–14.
- Acunzo M, Croce CM. MicroRNA in cancer and cachexia–a mini-review. J Infect Dis. 2015;212:S74–S77.
- Han YN, Xia SQ, Zhang YY, et al. Circular RNAs: a novel type of biomarker and genetic tools in cancer. Oncotarget 2017;8:64551–64563.
- Thyagarajan A, Shaban A, Sahu RP. MicroRNA-directed cancer therapies: implications in melanoma intervention. J Pharmacol Exp Ther. 2018;364:1–12.
- Cheng J, Chen Y, Zhao P, et al. Dysregulation of miR-638 in hepatocellular carcinoma and its clinical significance. Oncol Lett. 2017;13:3859–3865.
- Wang XX, Liu J, Tang YM, et al. MicroRNA-638 inhibits cell proliferation by targeting suppress PIM1 expression in human osteosarcoma. Tumour Biol: J Int Soc Oncodev Biol Med. 2016;37:16367–16375.
- Zhang J, Bian Z, Zhou J, et al. MicroRNA-638 inhibits cell proliferation by targeting phospholipase D1 in human gastric carcinoma. Protein Cell. 2015;6:680–688.
- Li M, Wang J, Liu H. Downregulation of miR-638 promotes progression of breast cancer and is associated with prognosis of breast cancer patients. Ott. 2018;11:6871–6877.
- Wei H, Zhang JJ, Tang QL. MiR-638 inhibits cervical cancer metastasis through Wnt/beta-catenin signalling pathway and correlates with prognosis of cervical cancer patients. Euro Rev Med Pharmacol Sci. 2017;21:5587–5593.
- Sciubba JJ, Larian B. Oral squamous cell carcinoma: early detection and improved 5-year survival in 102 patients. General Dentistry 2018;66:e11–e16.
- Prasad G, Seers C, Reynolds E, et al. A panel of microRNAs can be used to determine oral squamous cell carcinoma. J Oral Pathol Med: Off Public Int Assoc Oral Pathol Am Acad Oral Pathol. 2017;46:940–948.
- Shen Y, Chen H, Gao L, et al. MiR-638 acts as a tumour suppressor gene in gastric cancer. Oncotarget 2017;8:108170–108180.
- Shi M, Jiang Y, Yang L, et al. Decreased levels of serum exosomal miR-638 predict poor prognosis in hepatocellular carcinoma. J Cell Biochem. 2018;119:47116.
- Vishnoi A, Rani S. MiRNA biogenesis and regulation of diseases: an overview. Methods Mol Biol. 2017;1509:1–10.
- Brown HA, Thomas PG, Lindsley CW. Targeting phospholipase D in cancer, infection and neurodegenerative disorders. Nat Rev Drug Disc. 2017;16:351–367.
- Zhang Y, Frohman MA. Cellular and physiological roles for phospholipase D1 in cancer. J Biol Chem. 2014;289:22567–22574.
- Tang W, Liang R, Duan Y, et al. PLD1 overexpression promotes invasion and migration and function as a risk factor for Chinese glioma patients. Oncotarget 2017;8:57039–57046.
- Xiao J, Sun Q, Bei Y, et al. Therapeutic inhibition of phospholipase D1 suppresses hepatocellular carcinoma. Clin Sci (Lond). 2016;130:1125–1136.
- Gadiya M, Mori N, Cao MD, et al. Phospholipase D1 and choline kinase-alpha are interactive targets in breast cancer. Cancer Biol Ther. 2014;15:593–601.
- Kang DW, Lee SH, Yoon JW, et al. Phospholipase D1 drives a positive feedback loop to reinforce the Wnt/beta-catenin/TCF signalling axis. Cancer Res. 2010;70:4233–4242.
- Kang DW, Lee BH, Suh YA, et al. Phospholipase D1 inhibition linked to upregulation of ICAT blocks colorectal cancer growth hyperactivated by wnt/beta-catenin and PI3K/Akt signalling. Clin Cancer Res. 2017;23:7340–7350.
- Kang DW, Min do S. Positive feedback regulation between phospholipase D and Wnt signalling promotes Wnt-driven anchorage-independent growth of colorectal cancer cells. PloS One. 2010;5:e12109.
- Rheinwald JG, Beckett MA. Tumourigenic keratinocyte lines requiring anchorage and fibroblast support cultured from human squamous cell carcinomas. Cancer Res. 1981;41:1657–1663.