Abstract
Abnormal histone modifications have been recognized as an important contributing factor to the initiation and progression of osteosarcoma. Sirtuin 1 (Sirt1) up-regulation has been discovered in osteosarcoma cells. This study tested the influence of Sirt1 on histone H3 phosphorylation at threonine 3 (H3T3ph) in osteosarcoma cells, along with Sirt1-H3T3ph axis on osteosarcoma cell autophagy. Plasmids or si-RNAs transfection was carried out to alter Sirt1 or H3T3ph expressions. Co-immunoprecipitation analysis and GST pull-down assay were done to probe the relationship between Sirt1 and H3T3ph. Phosphoryltransferase activity of Sirt1 was tested by in vitro kinase activity assay. Cell autophagy was measured by a number of autophagosome, conversion of LC3-I to LC3-II, degradation of long-lived protein and ATG protein expressions. We found that Sirt1 directly interacted with H3 and phosphorylate H3T3 at threonine 3 in osteosarcoma cells. Moreover, Sirt1 facilitated osteosarcoma cell autophagy under starvation condition. H3T3ph took part in the Sirt1-facilitated osteosarcoma cell autophagy under starvation condition. Besides, Sirt1-H3T3ph axis facilitated osteosarcoma cell autophagy might be achieved through transcriptional activation of ATG genes. Sirt1 promoted osteosarcoma initiation and progression might be via phosphorylate H3T3 and then facilitate osteosarcoma cell autophagy through activating ATG genes transcription under starvation condition.
Introduction
Osteosarcoma is a frequently malignant bone sarcoma that usually happened in the first or second decade of life [Citation1]. Unfortunately, the long-term survival rate of patients with osteosarcoma has not significantly improved in recent years [Citation2]. Epigenetic modifications refer to modifications in the structure of DNA without alternations of DNA sequences, which are the results of DNA modification after replication and/or post-translational modification of proteins related to DNA [Citation3]. Aberrantly epigenetic modifications are demonstrated to be closely related to the onset and development of many human cancers, including osteosarcoma [Citation4,Citation5]. These modifications can influence multiple gene expressions and regulate many signaling pathways in tumor cells [Citation6]. It believes that further studying the biological mechanism of epigenetic modifications is of great significance for the therapy of osteosarcoma.
Histone modification is a key type of epigenetic modifications and regulatory mechanisms that affects the structure of chromatin [Citation7]. In general, histone can be phosphorylated, methylated, acetylated, ubiquitinated and ADP-ribosylated [Citation8]. Among them, phosphorylation, especially Histone H3 phosphorylation, is a common and important modification pathway, which is closely related to chromosome condensation during mitosis and transcription [Citation9]. Lee et al. reported that squamocin could regulate H3 phosphorylation and promoted G1 phase arrest and apoptosis of glioma, hepatocellular carcinoma and colon cancer cells [Citation10]. Espino et al. indicated that Ras-MAPK pathway-mediated H3 phosphorylation took part in the development of pancreatic cancer [Citation11]. Until now, no literature can be searched concerning the H3 phosphorylation in osteosarcoma. More experimental researches are still demanded to further probe whether H3 phosphorylation joins in the initiation and progression of osteosarcoma, along with the regulation of histone H3 phosphorylation.
Sirtuin 1 (Sirt1) is an extensively studied histone deacetylation and methylation enzyme in mammalian cells [Citation12]. It can influence multiple cellular biological processes, such as cell metabolism, inflammation, proliferation and DNA repair [Citation13]. Recent studies proved that Sirt1 also targeted some non-histone substrates in cells [Citation14,Citation15]. The up-regulation of Sirt1 has been discovered in many human cancer cells, including osteosarcoma cells [Citation16,Citation17]. It is demonstrated that Sirt1 can elevate osteosarcoma cell growth and metastasis [Citation18,Citation19].
Herein, we analyzed the influence of Sirt1 on H3 phosphorylation at threonine 3 (H3T3ph) in osteosarcoma cells. The regulatory activity of Sirt1-H3T3ph on osteosarcoma cell autophagy was also probed.
Materials and methods
Cell culture and treatment
WT Saos-2, MG-63, HOS cells were all received from Procell (Wuhan, China). Saos-2 cells were grown in McCoy’s 5 A medium (Procell) containing 15% fetal bovine serum (FBS, Hyclone, Logan, UT, USA) at 37 °C with 5% CO2. MG-63 and HOS cells were grown in MEM (Procell, Wuhan, China) containing 10% FBS at 37 °C with 5% CO2. Cells knock-in or knock-out Sirt1 and/or H3 genes were provided by Cyagen Biosciences (Taicang, China). To simulate starvation condition, cells were cultured in Earle’s balanced salt solution (EBSS, Gibco, CA, USA).
Plasmids or si-RNAs transfection
All plasmids used in our experiments were designed and synthesized by Taihe Biotechnology Co., Ltd (Beijing, China). siRNAs targeting Sirt1, ATG5, ATG13 and/or ATG14 were designed and synthesized by Thermo Scientific (MA, USA). Lipofectamine 3000 reagent (Invitrogen, CA, USA) was carried out for plasmids or siRNAs transfection. Culture medium containing 400 μg/ml neomycin (Sigma-Aldrich, MO, USA) was used for selecting stably transfected cells.
Co-immunoprecipitation analysis
Co-immuoprecipaitation analysis was carried out as earlier described [Citation20]. Briefly, followed by collection and sonication, cell lysate was mixed with mouse IgG or primary antibodies overnight. Subsequently, protein A/G agarose beads were supplemented into the lysates for 3 h. Finally, the precipitated proteins were released by boiling and analyzed by Western blotting.
Glutathione sepharose-transferase (GST) pull-down assay
GST pull-down assay was done as previously reported [Citation21]. Briefly, cells were gathered, rinsed using PBS, re-suspended in binding buffer and sonicated. Cell lysate was blended with GS-4B beads (GE Healthcare, Beijing, China) and spun down at 4 °C. Followed by rinsing, the beads were re-suspended in the binding buffer to produce 50% slurry. GST-Sirt1 was immobilized with this 50% bead slurry in binding buffer. After that, the beads were rotated for 1 h, rinsed, resuspended in binding buffer and reacted with in vitro expressed H3 protein. Finally, Western blotting was carried out to test binding proteins.
In vitro kinase activity assay
H3-containing nucleosomes were purified and in vitro kinase activity assay was done as previously described [Citation22]. For purification of H3-containing nucleosomes, cell nuclei were collected by Nuclear Extraction kit (Solarbio, Beijing, China). After digested using Mnase (Sigma-Aldrich), the nucleosomes were immunoprecipitated and resolved. For in vitro kinase activity assay, Sirt1 was reacted with nucleosomes containing 20 μM [γ-32P] ATP at 30 min. The phosphoproteins were tested by Western blotting and analyzed using Phosphor-Image Screens (Packard, Palo Alto, CA, USA).
Long-lived protein degradation assay
Long-lived protein degradation assay was done as previously described [Citation22]. Briefly, cells were labeled with 0.2 mCi/mL L-[14C] valine. Followed by growth in complete medium or EBSS medium supplemented with 6 mM valine for 5 h, trichloroacetic acid (TCA) was added into the culture medium to precipitate protein. After centrifugation, long-lived protein degradation was calculated by the ratio of acid-soluble radioactivity.
qRT-PCR
Total RNAs were separated by RNAiso Plus (Takara Biomedical Technology, Beijing, China). cDNA was synthesized with the help of PrimeScript cDNA systhesis kit (Takara Biomedical Technology). Then, the ATG5 and ATG7 mRNA levels were tested by TB Green Fast qPCR Mix (Takara Biomedical Technology) and compared to GAPDH expression. Results were calculated using 2-△△Ct method [Citation23].
Western blotting
All antibodies used in Western blotting were received from Abcam Biotechnology (MA, USA). Western blotting was carried out using Bio-Rad Bis-Tris Gel system (Bio-Rad Laboratories, CA, USA). The signals of proteins were recorded using Bio-Rad ChemiDocTM XRS system (Bio-Rad Laboratories) and Image LabTM software (Bio-Rad Laboratories).
Statistical analysis
Graphpad Prism V6.0 software (Graphpad Software, San Diego, CA, USA) was used for statistical analysis. Data were shown as mean ± standard deviation (SD). Differences between groups (p values) were computed using ANOVA. Significant difference was set at p ˂ .05.
Results
Sirt1 positively regulates H3T3ph in osteosarcoma cells
presented that there was a positive relationship between Sirt1 level and H3T3ph level in osteosarcoma cells. Saos-2 and MG-63 cells with high expression of Sirt1 also have high expression of H3T3ph, while HOS cells with low expression of Sirt1 havelow expression of H3T3ph. Exogenous Sirt1 (Sirt1+) was transfected into MG-63 and HOS cells to further test the positive relationship between Sirt1 and H3T3ph. Results in showed that Sirt1+ transfection significantly elevated the H3T3ph levels in MG-63 and HOS cells (p ˂ .001). Besides, two siRNAs targeting Sirt1 (si-Sirt1-1 and si-Sirt1-2) were also transfected into MG-63 and HOS cells. displayed that followed by si-Sirt1-1 or si-Sirt1-2 transfection, the H3T3ph levels in MG-63 and HOS cells were decreased. These outcomes indicated that Sirt1 could positively modulate H3T3ph in osteosarcoma cells.
Sirt1 directly interacts with H3 and phosphorylates H3T3 in osteosarcoma cells
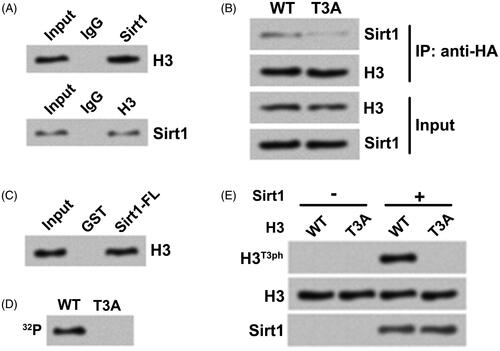
Then, MG-63 cell lysate was subjected to co-immunoprecipitation analysis with antibodies against Sirt1 and H3. pointed out that Sirt1 directly interacted with H3 in MG-63 cells and vice versa. To probe the relationship between this interaction and H3T3ph, HA-tagged WT H3-expressing plasmid and T3A-mutant (mimics de-phosphorylation of H3T3) H3-expressing plasmid were transfected into H3−/− (H3 knockout) HOS cells. showed that by contrast with H3WT, the concentration of Sirt1 bound to H3T3A was reduced, which suggested that H3T3ph was important to the association between Sirt1 and H3. presented that GST-tagged full-length Sirt1 directly bound to the HA-tagged H3WT. Besides, H3WT-containing nucleosomes and H3T3A-containing nucleosomes were reconstituted. The results of in vitro kinase activity assay in displayed that Sirt1 only phosphorylated H3WT-containing nucleosomes by using [γ-32P] as the phosphate donor. Western blotting of samples showed that the H3T3ph expression was only detected when Sirt1 and H3WT-containing nucleosomes co-existed (). These outcomes indicated that Sirt1 could directly interact with H3 and phosphorylate H3T3 in osteosarcoma cells.
Figure 1. Sirt1 positively regulates H3T3ph in osteosarcoma cells. (A) Western blotting was utilized for testing the protein levels of H3T3ph and Sirt1 in Saos-2, MG-63 and HOS cells. (B) Hexa-histidine (His6)-tagged Sirt1 (Sirt1+) plasmid was transfected into MG-63 and HOS cells. Western blotting was utilized for testing the protein levels of H3T3ph, H3 and Sirt1. (C) Two siRNAs targeting Sirt1 (si-Sirt1-1 and si-Sirt1-2) were transfected into MG-63 and HOS cells, respectively. Western blotting was utilized for testing the protein levels of H3T3ph, H3 and Sirt1. N = 3. ***p˂.001.
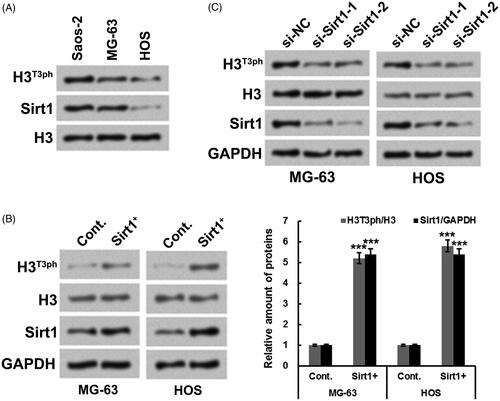
Figure 2. Sirt1 directly interacts with H3 and phosphorylates H3T3 in osteosarcoma cells. (A) The association between Sirt1 and H3 in MG-63 cells was tested by co-immunoprecipitation assay. (B) HA-tagged H3WT or H3T3A plasmid was transfected into H3−/− HOS cells. Co-immunoprecipitation assay was utilized to test Sirt1 binding. (C) GST pull-down assay was carried out to further test the relationship between Sirt1 and H3 using GST-tagged Sirt1. (D) Whether Sirt1 could phosphorylate H2B was detected using an in vitro kinase activity assay. (E) The samples from in vitro kinase activity assay were evaluated by Western blotting to measure the H3T3ph, H3 and Sirt1 levels. N = 3.
Sirt1 facilitates osteosarcoma cell autophagy in starvation condition
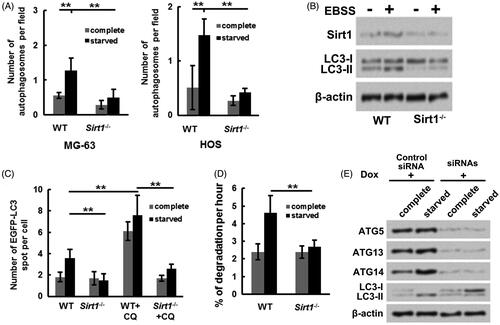
Further experiments were performed to probe the influence of Sirt1 on osteosarcoma cell autophagy under starvation condition. showed that relative to grown in complete medium, the number of autophagosomes in MG-63 and HOS cells were both increased grown in starved (EBSS) medium (p ˂ .01). However, the number of autophagosomes in Sirt1−/− MG-63 and HOS cells grown in starved medium were remarkably decreased (p ˂ .01). Moreover, displayed that the LC3-II/LC3-I expression rate in MG-63 cells was increased under starvation condition, while the LC3-II/LC3-I expression rate in Sirt1−/− MG-63 cells was not changed under complete medium or EBSS medium. These outcomes illustrated that Sirt1 might exert a key role in osteosarcoma cell autophagy. Besides, the plasmid expressing EGFP tagged LC3 was transfected into WT and Sirt1−/− HOS cells, EGFP-LC3 puncta was tested. presented that compared to WT HOS cells, the EGFP-LC3 puncta number under starvation condition was decreased in Sirt1−/− HOS cells (p ˂ .01). Chloroquine (CQ) was utilized as an autophagy inhibitor. The results showed that followed by CQ incubation, the EGFP-LC3 puncta number was only notably elevated in WT HOS cells (p ˂ .01), which implied that knockout of Sirt1 suppressed autophagosome formation. Besides, displayed that relative to WT HOS cells, the degradation of long-lived protein under starvation condition was repressed in Sirt1−/− HOS cells. An MG-63 cell line stably expressing Dox-inducible siRNA targeting ATG5, ATG13 and ATG14 was established. displayed that the increased ATG5, ATG13, ATG14 and LC3-II/LC3-I expression levels (rate) caused by Sirt1 overexpression and starvation stimulation were remarkably mitigated by Dox-inducible siRNA transfection. These above outcomes illustrated that Sirt1 could cause osteosarcoma cell autophagy under starvation condition via elevating core autophagy protein expressions.
Figure 3. Sirt1 facilitates osteosarcoma cell autophagy under starvation condition. Followed by maintained at complete medium or starved (EBSS) medium, (A) the numbers of autophagosomes in WT or Sirt1−/− MG-63 and HOS cells were counted, respectively, (B) the LC3-I and LC3-II levels in WT or Sirt1−/− MG-63 cells were measured. (C) Plasmid expressing EGFP-tagged LC3 was transfected into WT or Sirt1−/− HOS cells. Followed by maintainance at different conditions, the EGFP-LC3 puncta number was measured. (D) The degradation of long-lived protein in WT or Sirt1−/− HOS cells under complete or starvation condition were tested by long-lived protein degradation assay. (E) An MG-63 cell line stably expressing Dox-inducible siRNA targeting ATG5, ATG7 and ATG14 was constructed and transfected with Sirt1-expressing plasmid. Followed by maintained at different condition, Western blotting was carried out to test the protein levels of ATG5, ATG13, ATG14, LC3-I and LC3-II. N = 3. **p˂.01.
H3T3ph takes part in the Sirt1-facilitated osteosarcoma cell autophagy
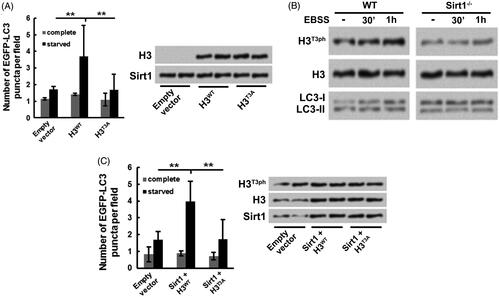
Subsequently, whether H3T3ph takes part in the Sirt1-caused osteosarcoma cell autophagy was analyzed. H3−/− Sirt1+/+ MG-63 cells were co-transfected with either H3WT or H3T3A and EGFP-LC3-expressing plasmids. showed that followed by starvation stimulation, the EGFP-LC3 puncta number in H3WT transfection group was significantly increased (p ˂ .01), while in H3T3A transfection group, the EGFP-LC3 puncta number was notably reduced (p ˂ .01). These results hinted that H3T3ph might take part in the Sirt1-caused osteosarcoma cell autophagy. presented that the H3T3ph level was increased in WT MG-63 cells with a time-dependent pathway after starvation stimulation, which was accompanied with the elevated expression of LC3-II/LC3-I. However, in Sirt1−/− MG-63 cells, the H3T3ph level was not changed after starvation stimulation. In addition, Sirt1 was overexpressed in WT MG-63 cells followed by starvation stimulation. displayed that the EGFP-LC3 puncta number was dramatically enhanced in Sirt1 + H3WT group (p ˂ .01) and reduced in Sirt1 + H3T3A group (p ˂ .01), which was accompanied with the changed expression of H3T3ph in MG-63 cells. These above outcomes illustrated that H3T3ph took part in the Sirt1-caused osteosarcoma cell autophagy.
Figure 4. H3T3ph takes part in the Sirt1-caused osteosarcoma cell autophagy. H3−/− Sirt1+/+ MG-63 cells were co-transfected with either H3WT or H3T3A and EGFP-LC3-expressing plasmids. Followed by growth at complete medium or starved medium, (A) the EGFP-LC3 puncta number, along with H3 and Sirt1 levels were detected, (B) the H3T3ph, LC3-I and LC3-II levels were measured. (C) Sirt1 was overexpressed in WT MG-63 cells. Followed by either H3WT or H3T3A and EGFP-LC3-expressing plasmids transfection and grown at different conditions, the EGFP-LC3 puncta number, along with H3T3ph, H3 and Sirt1 levels were measured. N = 3. **p˂.01.
Sirt1-H3T3ph axis facilitates osteosarcoma cell autophagy through activating ATG genes transcription
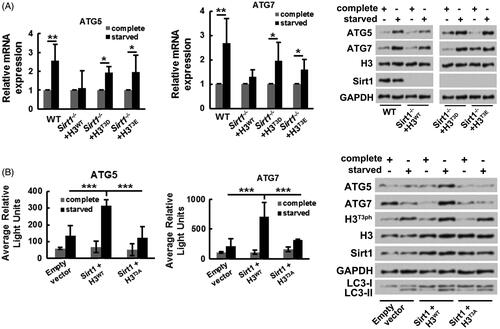
Finally, the influence of Sirt1-H3T3ph axis on transcriptional activities of ATG genes in osteosarcoma cells was explored. showed that followed by starvation stimulation, both the mRNA and protein expressions of ATG5 and ATG7 were increased in WT MG-63 cells (p ˂ .01 in mRNA level). However, followed by starvation stimulation, the increased mRNA and protein expressions of ATG5 and ATG7 were less in Sirt1−/− MG-63 cells transfected with H3WT, by contrast within Sirt1−/− MG-63 cells transfected with H3T3D or H3T3E (both mimics phosphorylation of H3T3, p ˂ .05 in mRNA level). These results implied that Sirt1-H3T3ph axis could elevate ATG genes expressions under starvation condition. In addition, displayed that Sirt1-H3T3ph axis notably promoted the transcriptional activities of ATG5 and ATG7 genes (p ˂ .001) under starvation condition, while H3T3A suppressed the transcriptional activities of ATG5 and ATG7 genes (p ˂ .001). The results of Western blotting pointed out that the expression rate of LC3-II/LC3-I was enhanced by transcriptional activation of ATG genes under starvation condition. These above outcomes illustrated that Sirt1 caused osteosarcoma cell autophagy might be achieved through phosphorylating H3T3 and then transcriptional activating of ATG genes.
Figure 5. Sirt1-H3T3ph axis facilitates osteosarcoma cell autophagy through activating ATG genes transcription. (A) WT or Sirt1−/− MG-63 cells were co-transfected with H3WT, H3T3D, or H3T3E. Followed by growth at different medium, the mRNA and protein expressions of ATG5 and ATG7 were tested by qRT-PCR and Western blotting, respectively. (B) MG-63 cells were co-transfected with Sirt1, H3WT or H3T3A-expressing plasmids and luciferase reporter plasmids that contain the promoter region of ATG5 and ATG7. Followed by growth at different medium, the transcriptional activities of ATG5 and ATG7 genes were recorded by average relative light unit using the Turner Designs Luminometer. The ATG5, ATG7, H3T3ph, H3, Sirt1, LC3-I and LC3-II levels were tested by Western blotting. N = 3. *p˂.05, **p˂.01 or ***p˂.001.
Discussion
Abnormal histone modifications have been recognized as important contributing factors to the initiation and progression of osteosarcoma [Citation4,Citation24,Citation25]. Herein, we discovered that Sirt1 could directly interact with H3 and phosphorylate H3T3 at threonine 3 in osteosarcoma cells. Moreover, we found that Sirt1 facilitated osteosarcoma cell autophagy in a starvation condition. H3T3ph took part in the Sirt1-facilitated osteosarcoma cell autophagy in starvation condition. Besides, we pointed out that Sirt1-H3T3ph axis facilitated osteosarcoma cell autophagy might be achieved through transcriptional activation of ATG genes.
Sirt1 is a member of the sirtuin family comprised of seven members, Sirt1 to Sirt 7 [Citation26]. As well-studied histone deacetylation and methylation enzyme, Sirt1 has been discovered to be highly expressed in a number of tumor cells, including osteosarcoma cells [Citation16,Citation26]. It is reported that overexpression of Sirt1 is associated with the activated oncogenes and silenced tumor suppressor genes in osteosarcoma cells [Citation27,Citation28]. H3 is a key component of nucleosome [Citation29]. Phosphorylation of H3 has been discovered to engage in the regulation of chromosome condensation and transcriptional modulation [Citation30]. Herein, we found that osteosarcoma cells with high expression of Sirt1 generally had high expression of H3T3ph. Overexpression of Sirt1 elevated the H3T3ph level in osteosarcoma cells, while the silence of Sirt1 lowered the H3T3ph level. The results of co-immunoprecipitation assay and GST pull-down assay indicated that Sirt1 could directly interact with H3 and phosphorylate H3T3. These outcomes illustrated that except for histone deacetylase and methylase activity, Sirt1 also could exhibit histone phosphoryl transferase activity.
Cell autophagy is an intracellular protein self-degradation process capable of maintaining cellular normal metabolism under nutrient shortage condition [Citation31]. Excessive activation of autophagy has been verified in many tumor cells, including osteosarcoma cells [Citation32]. Autophagy can offer tumor cells with a survival-promoting activity in response to the bad environment and tumor cells can activate autophagy to promote growth and aggressiveness [Citation33]. Sirt1 is found to engage in the modulation of a number of tumor cell autophagy [Citation34,Citation35]. Niu et al. reported that Sirt1 took part in the alisertib, an aurora kinase A inhibitor-activated osteosarcoma cell autophagy [Citation36]. In this research, we revealed that Sirt1 facilitated osteosarcoma cell autophagy in starvation condition. Absence of Sirt1 suppressed the generation of autophagosome, conversion of LC3-I to LC3-II, long-lived protein degradation and ATG protein expressions in osteosarcoma cells. What is more, we discovered that H3T3ph took part in the Sirt1-facilitated osteosarcoma cell autophagy in starvation condition. Followed by starvation stimulation, the H3T3ph level was increased in WT MG-63 cells, but not in Sirt1−/− MG-63 cells. These outcomes illustrated that Sirt1 facilitated osteosarcoma cell autophagy might be achieved via modulating H3T3ph.
The genes that join in cell autophagy process are named as ATG genes [Citation37]. ATG genes encode ATG proteins. Among them, ATG5 and ATG14 are closely related to the extension of autophagic vesicles; ATG7 is required for the conversion of LC3-I to LC3-II; ATG13 is important for the autophagosome initiation [Citation38]. In the current study, we discovered that the protein expressions of ATG5, ATG13 and ATG14 were all elevated in osteosarcoma cells under starvation condition. Moreover, Sirt1-H3T3ph axis enhanced the mRNA and protein expressions of ATG5 and ATG7 in osteosarcoma cells via activating ATG5 and ATG7 genes transcription under starvation condition. These outcomes indicated that Sirt1-H3T3ph axis facilitated osteosarcoma cell autophagy might be through activating ATG genes transcription.
Collectively, the influence of Sirt1 on H3T3ph in osteosarcoma cells and Sirt1-H3T3ph axis on osteosarcoma cell autophagy were investigated in this research. Sirt1 promoted osteosarcoma initiation and progression might be via phosphorylating H3T3 and then facilitating osteosarcoma cell autophagy through activating ATG genes transcription under starvation condition. More experiments in the future are still demanded to studying the spatial structure of Sirt1, which is of great importance for comprehending the histone phosphoryl transferase activity of Sirt1.
Acknowledgements
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Disclosure statement
The authors declare that they have no conflict of interest.
References
- Rogozhin DV, Bulycheva IV, Konovalov DM, et al. Classical osteosarcoma in children and adolescent. Arkh patol. 2015;77:68–74.
- Murakami T, Igarashi K, Kawaguchi K, et al. Tumor-targeting Salmonella typhimurium A1-R regresses an osteosarcoma in a patient-derived xenograft model resistant to a molecular-targeting drug. Oncotarget. 2017 ;8:8035–8042.
- Kanwal R, Gupta K., Gupta S. Cancer epigenetics: an introduction In: Verma M, editor. Cancer epigenetics: risk assessment, diagnosis, treatment, and prognosis. New York, NY: Springer New York; 2015. p. 3–25.
- Morrow JJ, Khanna C. Osteosarcoma genetics and epigenetics: emerging biology and candidate therapies. Crit Rev Oncog. 2015;20:173–197.
- Kinnaird A, Zhao S, Wellen KE, et al. Metabolic control of epigenetics in cancer. Nat Rev Cancer.. 2016;16:694.
- Jones PA, Issa J-PJ, Baylin S. Targeting the cancer epigenome for therapy. Nat Rev Genet. 2016;17:630.
- Wang R, Xin M, Li Y, et al. The Functions of Histone Modification Enzymes in Cancer. CPPS. 2016;17:438–445.
- Shanmugam MK, Arfuso F, Arumugam S, et al. Role of novel histone modifications in cancer. Oncotarget. 2018;9:11414–11426.
- Hans F, Dimitrov S. Histone H3 phosphorylation and cell division. Oncogene. 2001;20:3021–3027.
- Lee CC, Lin YH, Chang WH, et al. Squamocin modulates histone H3 phosphorylation levels and induces G1 phase arrest and apoptosis in cancer cells. BMC cancer. 2011;11:58.
- Espino PS, Pritchard S, Heng HH, et al. Genomic instability and histone H3 phosphorylation induction by the Ras-mitogen activated protein kinase pathway in pancreatic cancer cells. Int J Cancer. 2009;124:562–567.
- McAndrews KM, LeBleu VS, Kalluri R. SIRT1 regulates lysosome function and exosome secretion. Dev Cell. 2019;49:302–303.
- Yang H, Bi Y, Xue L, et al. Multifaceted modulation of SIRT1 in cancer and inflammation. Crit Rev Oncog.. 2015;20:49–64.
- Vaziri H, Dessain SK, Ng Eaton E, et al. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159.
- Yeung F, Hoberg JE, Ramsey CS, et al. Modulation of NF-kappaB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 2004;23:2369–2380.
- Zhang N, Xie T, Xian M, et al. SIRT1 promotes metastasis of human osteosarcoma cells. Oncotarget. 2016;7:79654–79669.
- Chen J, Cao L, Li Z, et al. SIRT1 promotes GLUT1 expression and bladder cancer progression via regulation of glucose uptake. Hum Cell. 2019 ;32:193–201.
- Yu XJ, Guo XZ, Li C, et al. SIRT1-ZEB1-positive feedback promotes epithelial-mesenchymal transition process and metastasis of osteosarcoma. J Cell Biochem. 2019;120:3727–3735.
- He S, Wang Z, Tang H, et al. MiR-217 inhibits proliferation, migration, and invasion by targeting SIRT1 in osteosarcoma. Cancer Biother Radiopharm. 2019;34:264–270.
- Luo H, Shenoy AK, Li X, et al. MOF acetylates the histone demethylase LSD1 to suppress epithelial-to-mesenchymal transition. Cell reports. 2016;15:2665–2678.
- Machida K, Liu B. Binding assays using recombinant SH2 domains: far-Western, pull-down, and fluorescence polarization. Methods Mol Biol. 2017;1555:307–330.
- Liu Y, Long YH, Wang SQ, et al. JMJD6 regulates histone H2A.X phosphorylation and promotes autophagy in triple-negative breast cancer cells via a novel tyrosine kinase activity. Oncogene. 2019 ;38:980–997.
- Ish-Shalom S, Lichter A. Analysis of fungal gene expression by real time quantitative PCR. Methods Mol Biol. 2010;638:103–114.
- La Noce M, Paino F, Mele L, et al. HDAC2 depletion promotes osteosarcoma's stemness both in vitro and in vivo: a study on a putative new target for CSCs directed therapy. J Exp Clin Cancer Res. 2018;37:296.
- He C, Liu C, Wang L, et al. Histone methyltransferase NSD2 regulates apoptosis and chemosensitivity in osteosarcoma. Cell death Dis. 2019;10:65.
- Liu T, Liu PY, Marshall GM. The critical role of the class III histone deacetylase SIRT1 in cancer. Cancer Res. 2009;69:1702–1705.
- Feng H, Guo P, Wang J, et al. Expression of Leptin and Sirtuin-1 is associated with poor prognosis in patients with osteosarcoma. Pathol Res Pract. 2016;212:319–324.
- Jia D, Niu Y, Li D, et al. lncRNA C2dat1 promotes cell proliferation, migration, and invasion by targeting miR-34a-5p in osteosarcoma cells. Oncol Res. 2018;26:753–764.
- Fyodorov DV, Zhou BR, Skoultchi AI, et al. Emerging roles of linker histones in regulating chromatin structure and function. Nat Rev Mol Cell Biol. 2018;19:192–206.
- Nowak SJ, Corces VG. Phosphorylation of histone H3: a balancing act between chromosome condensation and transcriptional activation. TIG. 2004;20:214–220.
- Bhat P, Kriel J, Shubha Priya B, et al. Modulating autophagy in cancer therapy: advancements and challenges for cancer cell death sensitization. Biochemical pharmacology. 2018;147:170–182.
- Lozy F, Karantza V. Autophagy and cancer cell metabolism. Semin Cell Dev Biol. 2012;23:395–401.
- White E. The role for autophagy in cancer. J Clin Invest. 2015;125:42–46.
- Xiong H, Ni Z, He J, et al. LncRNA HULC triggers autophagy via stabilizing Sirt1 and attenuates the chemosensitivity of HCC cells. Oncogene. 2017;36:3528.
- Wang J, Li J, Cao N, et al. Resveratrol, an activator of SIRT1, induces protective autophagy in non-small-cell lung cancer via inhibiting Akt/mTOR and activating p38-MAPK. OTT. 2018;11:7777–7786.
- Niu NK, Wang ZL, Pan ST, et al. Pro-apoptotic and pro-autophagic effects of the Aurora kinase A inhibitor alisertib (MLN8237) on human osteosarcoma U-2 OS and MG-63 cells through the activation of mitochondria-mediated pathway and inhibition of p38 MAPK/PI3K/Akt/mTOR signaling pathway. Drug Des Dev Ther. 2015;9:1555–1584.
- Levine B, Kroemer G. Biological functions of autophagy genes: a disease perspective. Cell. 2019;176:11–42.
- Dikic I, Elazar Z. Mechanism and medical implications of mammalian autophagy. Nat Rev Mol Cell Biol. 2018;19:349–364.
