Abstract
Objective
The long intergenic non-coding RNA 01296 (LINC01296) has been reported to be overexpressed in multiple tumours. However, the role of LINC01296 in clinicopathologic and prognostic value in cancers remains completely unknown. The aim of the present meta-analysis was to comprehensively elucidate the correlation between LINC01296 with clinicopathological features and survival outcomes in tumours.
Methods
Electronic databases of PubMed, Web of Science, Chinese National Knowledge Infrastructure (CNKI), and Wanfang Database were used to search relevant studies. The role of LINC01296 in cancers was evaluated by pooled hazard ratios (HRs), odds ratios (ORs) and 95% confidence intervals (CIs).
Results
In total, nine studies compromising 720 participants were enrolled in this analysis. The pooled results showed increased LINC01296 expression could predict unfavourable overall survival (OS) (HR = 1.89, 95%CI = 1.47–2.43, p < .001). Additionally, elevated LINC01296 expression was correlated with clinical stage (OR = 2.95, 95%CI = 2.13–4.08, p < .001), lymph node metastasis (OR = 2.76, 95%CI = 2.00–3.81, p < .001), tumour size (OR = 2.80, 95%CI = 1.77–4.41, p < .001), and tumour differentiation (OR = 2.11, 95%CI = 1.36–3.27, p < .001) in patients with cancers.
Conclusion
The results of this meta-analysis indicated LINC01296 was a novel biomarker for prognosis and clinicopathological parameters in cancers.
Introduction
Cancer is becoming the leading public health problem throughout the world. In 2018, there were a predicted 18.1 million new cases and 9.6 million deaths of cancers worldwide based on a report by the International Agency for Research on Cancer [Citation1]. Despite major advances in the treatment of tumours over the last decade, the 5-year survival rate for tumours remained worse, mainly owing to no ideal biomarkers for the early detection and effective treatment of tumours. Thus, it is urgent to explore a promising biological target in precise therapy and prognostication of cancer.
Long non-coding RNAs (lncRNAs) belong to the typical class of non-coding RNAs (ncRNAs) comprising a transcription length of more than 200 nucleotides [Citation2,Citation3]. Emerging studies have demonstrated lncRNAs play vital roles in the physiological and pathological process of cancers, such as cell proliferation, migration, differentiation, metastasis, and invasion, by functioning as oncogene or tumour suppressor [Citation4,Citation5]. In addition, accumulating evidence has shown that lncRNAs can be recognized as tumour-specific prognostic predictors for some cancers [Citation3]. Recent meta-analyses have suggested that lncRNA HOTAIR [Citation6], PVT1 [Citation7], ZEB1-AS1 [Citation8], BCRA4 [Citation9] and CASC2 [Citation10] are associated with clinicopathologic characteristics and prognosis in various cancers, and lncRNAs may be candidates for predicting biomarkers for precise cancer prognosis.
LINC01296, known as Long intergenic non-coding RNA 01296, has attracted great interest as the abnormal expression of LINC01296 has been reported in different human tumours [Citation11,Citation12]. Previous studies have demonstrated that LINC01296 is upregulated in kinds of cancers, including gastric cancer, breast cancer, colorectal cancer, pancreatic cancer, and no small cell lung cancer. It has been shown to contribute to tumour proliferation, invasion, and migration [Citation4,Citation13–15]. Furthermore, high expression of LINC01296 level has been found to be positively correlated to clinicopathologic features with LINC01296 overexpression exhibiting worse overall survival (OS), low differentiation, high tumour node metastasis (TNM) stage, and lymph node metastasis (LNM) [Citation4,Citation14,Citation16,Citation17]. However, owing to the discrete outcomes and limited sample size in current studies, we carried out this meta-analysis to elucidate the association of LINC01296 between the outcome and clinicopathologic characteristics, thus, examining the potential value of LINC01296 as a promising predictor in human cancers.
Materials and methods
Literature searching strategies
We searched several electronic databases of PubMed, Web of Science, Chinese National Knowledge Infrastructure (CNKI), and Wanfang database. The keywords were as follows: “LINC01296” AND “Tumour” OR “Cancer” OR “Carcinoma” AND “Survival” OR “Outcome” OR “Predict”. The latest search ended on March 2019.
Inclusion and exclusion criteria
Inclusion criteria were as follows: (1) study reporting clinical role of LINC01296 with human tumours; (2) study has assessed the correlation of LINC01296 levels with prognosis or clinicopathological parameters; (3) study showing patients with high and low expression of LINC01296 levels in tumour tissues; (4) study providing available data for extraction or calculation of hazard ratios (HRs) and 95% confidence intervals (CIs) for OS. Exclusion criteria were the following: (1) reviews, letters, conference reports and animal studies; (2) studies lacking complete data to calculate the HRs, 95%CI, and p-values.
Data extraction and quality assessment
Two investigators (C. Z., and W. S.) independently examined all literature based on the inclusion and exclusion criteria, and any discrepancies were discussed to an agreement. The following information were carefully extracted: author, year, country, type of cancer, sample size, cut-off value, number of patients in different LINC01296 expression groups, number of patients with TNM stage, LNM, tumour differentiation in different expression groups, detection method, follow-up months, outcome (OS and DFS), and HRs and 95% CIs. If HRs and 95% CIs were analyzed by multivariate and univariate analysis, the former was the priority. If they could not be obtained, Engauge Digitizer Version 10.4 (http://markummitchell.github.io/engauge-digitizer/) and published method [Citation18] were performed to extract survival data through Kaplan–Meier curves in publications. Study quality was evaluated by the Newcastle–Ottawa quality assessment Scale (NOS). Score items were classified into three major categories: selection, assessment of outcome, and comparability. If the score ≥ 7, the quality of the study was considered as high.
Statistical analysis
All extracted data were analyzed in this meta-analysis by using STATA software version 12.0 (StataCorp LLC, College Station, TX, USA). Pooled HRs and 95% CIs were calculated to assess the correlation of LINC01296 expression with prognosis in cancers. If observed HR > 1, it implied that patients with high LINC01296 expression had a worse clinical outcome. Pooled odds ratios (ORs) and 95% CIs were used to evaluate the association of LINC01296 expression and clinicopathological features. The heterogeneity among studies was performed by Chi-squared test and I2 statistics. If the heterogeneity was not significant (p > .05 for Chi-squared test or I2 < 50%), we used the fixed-effect model. Otherwise, the random-effect model was chosen [Citation19]. Subgroup analysis stratified by cancer type, sample size, follow-up months and HR estimation method were conducted to analyze sources of heterogeneity. Sensitivity analysis was applied to examine the stability of the results. Furthermore, publication bias was assessed by funnel plots and Begg’s test. p-Values < .05 meant statistically significant.
Results
Study identification and characteristics
A total of nine eligible studies were enrolled in this analysis ultimately [Citation4,Citation12,Citation13,Citation15–17,Citation20–22]. The flow diagram was shown in . All studies were published from 2017 to 2019. There were totally 720 patients with sample size ranging between 40 and 221. According to the results measured by qRT-PCR, all patients were divided into two groups: high and low LINC01296 expression group. Among the enrolled studies, there were 8 different kinds of tumour: gastric cancer, oesophagal squamous cancer, pancreatic adenocarcinoma, cholangiocarcinoma, bladder cancer, prostate cancer, non-small cell lung cancer, and breast cancer. The major characteristics of eligible articles were summarized in .
Table 1. Characteristics of the eligible studies included in the meta-analysis (n = 9).
Association of LINC01296 and OS
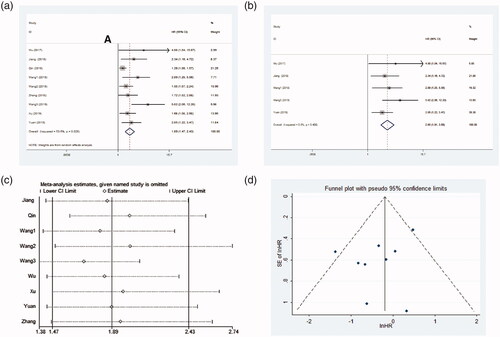
Nine studies including 720 patients assessed the HR and 95% CI of OS. Because significant heterogeneity was observed between studies (I2 = 53.6%, p = .028, ), a random-effect model was conducted. As indicated in , the pooled HR indicated that patients with high LINC01296 level significantly had a worse prognosis (pooled HR = 1.89, 95%CI = 1.47–2.43, p < .001). To explore underlying sources of heterogeneity, subsequent subgroup analyses were performed by tumour type, sample capacity, follow-up period, and HR obtained measurements (). Subgroup analysis showed LINC01296 overexpression could estimate unfavourable outcome in digestive system (HR = 1.62, 95%CI = 1.27–2.06, p < .001; I2 = 41.5%, p = .145), and other systems (HR = 2.56, 95%CI = 1.53–4.28, p < .001; I2 = 45.7%, p = .137) (). After stratified by sample size, a significant relationship between the high LINC01296 expression and an unfavourable prognosis of patients in studies with sample size <70 (HR = 1.85, 95%CI = 1.29–2.65, p = .001; I2 = 64.9%, p = .022) and ≥70 (HR = 1.96, 95%CI = 1.42–2.71, p < .001; I2 = 17.1%, p = .306) were observed (). In addition, the increased LINC01296 expression predicted a worse OS in studies with follow-up ≥60 months (HR = 1.79, 95%CI = 1.38–2.31, p < .001; I2 = 43.4%, p = .102), while there was no significant difference between LINC01296 overexpression and OS in studies with follow-up <60 months (HR = 2.59, 95%CI = 0.83–8.14, p = .102; I2 = 82.5%, p = .017) (). After stratified analysis by HR estimation method, the pooled results demonstrated a statistically significant connection in studies with indirect method subgroup (HR = 1.41, 95% CI = 1.21–1.65; I2=0.0%, p = .529) and direct method subgroup (HR = 2.65, 95% CI = 1.91–3.68; I2=0.0%, p = .488) (). Taken together, these results indicated that tumour type (digestive system vs other systems) and estimate method (indirectly vs directly estimation method) mainly explained the sources of heterogeneity.
Independent prognostic value of LINC01296
Five studies with 485 patients assessed the association between LINC01296 expression and OS in a Cox proportional multivariable model. The pooled results demonstrated that elevated LINC01296 expression experienced a higher risk of shorter OS, suggesting that LINC01296 functioned as an independent biomarker for tumour prognosis (HR = 2.65, 95%CI = 1.91–3.68, p < .000, ). No significant heterogeneity was found between the studies (I2 = 0.0%, p = .488).
Correlation between LINC01296 and clinicopathologic characteristics
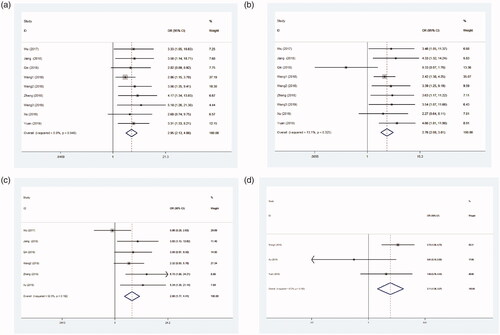
High LINC01296 expression level was observed to be significantly related with poor TNM (III + IV vs. I + II, OR = 2.95, 95% CI = 2.13–4.08, p < .001, ), LNM (yes vs. no, OR = 2.76, 95% CI = 2.00–3.81, p < .001, ), tumour size (large vs. small, OR = 2.80, 95% CI = 1.77–4.41, p < .001, ), and low tumour differentiation (low vs. high, OR = 2.11, 95% CI = 1.36–3.27, p = .001, ).
Sensitivity analysis and publication bias
After removing each study, the pooled HR was not significantly changed, showing that the results were stable (). The shape of funnel plots presented absence of asymmetry (). Similarly, Begg’s test indicated no obvious evidence for publication bias (p = .211).
Discussion
Accumulating studies have shown that aberrant expression of lncRNAs plays important roles in the progression of multiple cancers [Citation23–25]. In recent years, LINC01296 has been reported to be upregulated in many cancers, including non-small cell lung cancer, urinary system cancers, gastric cancers and digestive tract tumours [Citation14–16,Citation21,Citation22]. Here we conducted this meta-analysis to evaluate the relationship of LINC01296 with clinicopathological parameters and prognostic value in different cancers. Interestingly, our results suggested that LINC01296 could function as a useful biomarker for tumours.
A total of nine eligible studies with 720 cancer patients, reporting eight common cancer types, finally met inclusion criteria. Our results demonstrated that increased LINC01296 level was significantly correlated with poor survival rates in kinds of cancers. Then, we systematically assessed the relationship between elevated LINC01296 with clinicopathological parameters. Interestingly, LINC01296 overexpression has been revealed to be significantly associated with clinical stage, LNM, tumour size and tumour differentiation, suggesting that elevated LINC01296 expression had a connection with advanced cancer features. Collectively, these observations provide evidence that LINC01296 acts as a novel prognostic target in cancers.
However, the potential molecular mechanisms underlying correlation of aberrant LINC01296 expression and poor clinical prognosis in cancers remained understood. Previous studies have demonstrated that LINC01296 induces cell proliferation and mediates cell migration through regulating gene activity in tumours [Citation4,Citation5,Citation13,Citation20,Citation26,Citation27]. It has been indicated that LINC01296 may serve as a competing endogenous RNA (ceRNA) to suppress miRNAs functions. In colorectal cancer, Liu et al have found that upregulation of LINC01296 contributes to cancer progression by LINC01296/miR-26a/GALNT3 axis [Citation5]. In cholangiocarcinoma, Zhang et al. have demonstrated that LINC01296 functions as a ceRNA for miR-5095 to induce tumour growth and progression [Citation21]. Yuan et al. have shown that LINC01296 promotes cell proliferation by regulating the Bcl-2/caspase-3 pathway in pancreatic adenocarcinoma [Citation20]. LINC01296 has been found to be an oncogenic lncRNA that participates in gastric cancer cells progression by sponging to miR-122, thus to affect MMP-9 regulatory pathway [Citation13]. In oesophagal squamous cell carcinoma, Wang et al. have reported that knockdown of LINC01296 inhibits cell growth via upregulating KLF2 expression in vivo [Citation12]. In bladder cancer, it has been found that LINC0296 aggregates cancer cell proliferation and invasion through affecting epithelial-mesenchymal transition (EMT) [Citation16]. In prostate cancer, Wu et al. have revealed that LINC01296 stimulates cell growth by effecting PI3K/Akt/mTOR signalling and EMT [Citation17]. In osteosarcoma, Yu et al. have reported that LINC01296 contributes to the proliferation, metastasis and cell cycle via cyclinD1 [Citation27]. In addition, Xu et al. have shown that LINC01296 promotes tumourigenesis of NSCLC by LINC01296/miR-598/Twist1 axis [Citation15]. Therefore, these pieces of evidence suggest that LINC01296 plays key roles in tumour progression and may be a candidate target for cancer prognosis.
Nevertheless, several limitations existed in this meta-analysis. Firstly, we could not pool results by one single type of tumour because of the relatively small sample size. Thus, we evaluated the role of LINC01296 expression by dividing into the digestive system and nondigestive system cancers to confirm the prognostic value of LINC01296. Secondly, all included studies were from China, making it more suitable for the Chinese or the Asian population context. Large-scale and well-designed studies are still needed to verify the clinical value of LINC01296 in different ethnicities. Thirdly, some of the HRs and 95%CI were calculated indirectly. Therefore, the clinical significance of increased LINC01296 expression in various cancers might be overestimated.
In conclusion, this meta-analysis indicates that LINC01296 overexpression correlates with survival outcomes and clinical parameters of cancers. In future, further well-designed and larger sample size studies are needed to validate the clinical value of LINC01296 in different cancers of various ethnic populations.
Author contributions
W. F. designed the study, analyzed the data and wrote the manuscript; C. Z., and W. S. collected the data; Q. Z. and X. Y. participated in analyzing the data; J. W. and Q. W. took part in writing the manuscript; M. L. assisted in study design and revised the manuscript; All the authors have reviewed and approved the final proof.
Acknowledgements
The authors would thank Dr. Yang Song and Xiao Meng of Pfizer Medical for helpful suggestions.
Disclosure statement
All authors have declared there are no conflicts of interest.
Additional information
Funding
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424.
- Weidle UH, Birzele F, Kollmorgen G, et al. Long Non-coding RNAs and their role in metastasis. Cancer Genomics Proteomics. 2017;14:143–160.
- Yarmishyn AA, Kurochkin IV. Long noncoding RNAs: a potential novel class of cancer biomarkers. Front Genet. 2015;6:145–145.
- Jiang M, Xiao Y, Liu D, et al. Overexpression of long noncoding RNA LINC01296 indicates an unfavourable prognosis and promotes tumourigenesis in breast cancer. Gene. 2018;675:217–224.
- Liu B, Pan S, Xiao Y, et al. LINC01296/miR-26a/GALNT3 axis contributes to colorectal cancer progression by regulating O-glycosylated MUC1 via PI3K/AKT pathway. J Exp Clin Cancer Res.. 2018;37:316.
- Abdeahad H, Avan A, Pashirzad M, et al. The prognostic potential of long noncoding RNA HOTAIR expression in human digestive system carcinomas: a meta-analysis. J Cell Physiol. 2019;234:10926–10933.
- Xiao M, Feng Y, Liu C, et al. Prognostic values of long noncoding RNA PVT1 in various carcinomas: an updated systematic review and meta-analysis. Cell Prolif. 2018;51:e12519.
- Wu Y, Ding M, Wei S, et al. The prognostic value of long noncoding RNA ZEB1-AS1 on clinical outcomes in human cancer. J Cancer. 2018;9:3690–3698.
- Zhao W, Wang Z, Fang X, et al. Long noncoding RNA Breast cancer antiestrogen resistance 4 is associated with cancer progression and its significant prognostic value. J Cell Physiol. 2018;234:12956–12963.
- Cai J, Zuo X, Chen Z, et al. Prognostic value and clinical significance of long noncoding RNA CASC2 in human malignancies: a meta-analysis. CMAR. 2018;10:1403–1412.
- Seitz AK, Christensen LL, Christensen E, et al. Profiling of long non-coding RNAs identifies LINC00958 and LINC01296 as candidate oncogenes in bladder cancer. Sci Rep. 2017;7:395.
- Wang L, Meng D, Wang Y, et al. Long non-coding RNA LINC01296 promotes esophageal squamous cell carcinoma cell proliferation and invasion by epigenetic suppression of KLF2. Am J Cancer Res. 2018;8:2020–2029.
- Qin QH, Yin ZQ, Li Y, et al. Long intergenic noncoding RNA 01296 aggravates gastric cancer cells progress through miR-122/MMP-9. Biomed Pharmacother. 2018;97:450–457.
- Qiu JJ, Yan JB. Long non-coding RNA LINC01296 is a potential prognostic biomarker in patients with colorectal cancer. Tumour Biol. 2015;36:7175–7183.
- Xu L, Wei B, Hui H, et al. Positive feedback loop of lncRNA LINC01296/miR-598/Twist1 promotes non-small cell lung cancer tumourigenesis. J Cell Physiol. 2019;234:4563–4571.
- Wang X, Wang L, Gong Y, et al. Long noncoding RNA LINC01296 promotes cancer-cell proliferation and metastasis in urothelial carcinoma of the bladder. Onco Targets Ther. 2019;12:75–85.
- Wu J, Cheng G, Zhang C, et al. Long noncoding RNA LINC01296 is associated with poor prognosis in prostate cancer and promotes cancer-cell proliferation and metastasis. OTT. 2017;10:1843–1852.
- Tierney JF, Stewart LA, Ghersi D, et al. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8:16.
- Liang J, Wei X, Liu Z, et al. Long noncoding RNA CYTOR in cancer: a TCGA data review. Clin Chim Acta. 2018;483:227–233.
- Yuan Q, Zhang Y, Feng L, et al. Upregulated long noncoding RNA LINC01296 indicates a dismal prognosis for pancreatic ductal adenocarcinoma and promotes cell metastatic properties by affecting. J Cell Biochem. 2019;120:552–561.
- Zhang D, Li H, Xie J, et al. Long noncoding RNA LINC01296 promotes tumour growth and progression by sponging miR-5095 in human cholangiocarcinoma. Int J Oncol. 2018;52:1777–1786.
- Wang B, Liang T, Li J. Long noncoding RNA LINC01296 is associated with poor prognosis in ESCC and promotes ESCC cell proliferation, migration and invasion. Eur Rev Med Pharmacol Sci. 2018;22:4524–4531.
- Shen X, Bai Y, Luo B, et al. Upregulation of lncRNA BANCR associated with the lymph node metastasis and poor prognosis in colorectal cancer. Biol Res. 2017;50:32.
- Gao C, Li H, Zhuang J, et al. The construction and analysis of ceRNA networks in invasive breast cancer: a study based on The Cancer Genome Atlas. CMAR. 2018;11:1–11.
- Wang L, Chunyan Q, Zhou Y, et al. BCAR4 increase cisplatin resistance and predicted poor survival in gastric cancer patients. Eur Rev Med Pharmacol Sci. 2017;21:4064–4070.
- Feng L, Houck JR, Lohavanichbutr P, et al. Transcriptome analysis reveals differentially expressed lncRNAs between oral squamous cell carcinoma and healthy oral mucosa. Oncotarget. 2017;8:31521–31531.
- Yu X, Pang L, Yang T, et al. lncRNA LINC01296 regulates the proliferation, metastasis and cell cycle of osteosarcoma through cyclin D1. Oncol Rep. 2018;40:2507–2514.