Abstract
Long non-coding RNAs (LncRNAs) have been proven to be involved in the progression of osteosarcoma. LncRNA DLEU1 was found to act as an oncogene in various types of cancer. In this study, we aimed to explore the biological functions of DLEU1 in osteosarcoma and the specific mechanism. Our results showed that DLEU1 was highly expressed in osteosarcoma tissue specimens and cells. Downregulation of DLEU1 in osteosarcoma cells inhibited the cell proliferation, migration and invasion. Further mechanistic analysis and functional assays demonstrated that DLEU1 exerted its role by sponging microRNA (miR)-671-5p in osteosarcoma cells. Moreover, DEAD-box helicase 5 (DDX5) was confirmed as the target of miR-671-5p. Furthermore, rescue assays indicated that miR-671-5p inhibitor significantly reversed the effects of DLEU1 knockdown on osteosarcoma cell proliferation and invasion while restoration of DDX5 rescued the proliferation and invasion of osteosarcoma cells transfected with miR-671-5p mimics. In conclusion, DLEU1 acted as an oncogene in osteosarcoma by directly sponging miR-671-5p and regulating the expression of DDX5. These findings suggested that DLEU1 might be a novel therapeutic target in the treatment of osteosarcoma.
Introduction
Osteosarcoma is the most common primary malignant tumour of bone with high morbidity in infants and adolescents [Citation1]. In recent years, exploring the molecular mechanism of osteosarcoma has gained considerable attention; however, the mechanisms underlying its initiation and progression have not been fully understood. Long non-coding RNAs (lncRNAs) are a group of non-protein coding transcripts with a length about 200 nucleotides [Citation2]. Recent studies have discovered that lncRNAs are involved in the initiation, growth, and metastasis of osteosarcoma [Citation3,Citation4].
An increasing number of researchers set to discover associated lncRNAs in the pathogenesis of osteosarcoma. Li et al. [Citation5] firstly reported that 403 lncRNAs are upregulated and 798 lncRNAs are downregulated in osteosarcoma tissues as compared to paired adjacent non-tumour tissues. Increasingly, many lncRNAs have been reported to play important roles in multiply biological processes of osteosarcoma including tumour cell proliferation, invasion, migration, and chemoresistance [Citation3,Citation4]. LncRNA DLEU1 is overexpressed and exerts an oncogene function by regulating cell proliferation, migration and invasion in several types of cancers including bladder cancer [Citation6], cervical cancer [Citation7], oral squamous cell carcinoma [Citation8], non-small cell lung cancer [Citation9], and colorectal cancer [Citation10]. However, the role of DLEU1 in osteosarcoma remains unclear.
It has been well established that lncRNAs can act as molecular sponges, scaffolds, and guides to interact with proteins, mRNAs, or miRNAs, thereby regulating the expression of target genes [Citation11,Citation12]. Growing evidence indicates that the interactions between lncRNAs and miRNAs play crucial roles in modulating tumour progression [Citation13]. In the current study, we investigated the role of DLEU1 in osteosarcoma and explored the interaction between DLEU1 and its target miRNA in vitro. The results indicated that DLEU1 acted as an oncogene in osteosarcoma by sponging miR-671-5p and regulating its downstream target gene DDX5 ().
Materials and methods
Clinical tissues and cell culture
Fifty paired osteosarcoma tissues and adjacent non-tumour tissues were obtained from Huaihe Hospital of Henan University (Kaifeng, China). All the tissue specimens were collected from osteosarcoma patients underwent surgical resection. The clinical samples were stored at −80 °C. The Ethics Committee of Huaihe Hospital of Henan University reviewed and approved the use of clinical tissues samples and the procedure of the study.
Human normal osteoblast hFOB1.19 cells and four osteosarcoma cell lines including HOS, MG63, U2OS, and Saos-2 cells were obtained from the Bank of Type Culture Collection of Chinese Academy of Sciences (Shanghai, China). The cells were cultured in either F12/DMEM or DMEM medium with 10% fetal bovine serum (FBS) and maintained in a 5% CO2 incubator at 37 °C.
Cell transfection
The expression of DLEU1 in Saos-2 cells was suppressed by transfection with siRNA-mediated silencing. Specific siRNA for DLEU1 (si-DLEU1) and its non-targeting control (si-control) obtained from GenePharma (Shanghai, China) were transfected into Saos-2 cells using Lipofectamine RNAimax (Invitrogen, Carlsbad, CA, USA).
Saos-2 cells were cultured in 6-well plates and then transfected with miR-671-5p mimics or DDX5 plasmid and their corresponding controls (miR-NC or vector) using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer’s instructions. After 48 h, cells were collected for the following experiments.
Cell proliferation assay
Saos-2 cells (5 × 103 cells/well) with different transfections were seeded in 96-well plates and cultured for 0, 24, 48, 72 h. Then the cells were added with 10 μl CCK-8 solution (Dojindo Molecular Technologies, Kyushu, Japan). Four hours later, OD value at the wavelength of 450 nm was measured using a Microplate Reader (Bio-Rad, Hercules, CA, USA).
Cell migration and invasion assay
Transwell assay was performed using transwell chambers (8 μm, Corning Inc., Corning, NY, USA). For the determination of cell invasion, the transwell inserts were coated with Matrigel (BD Biosciences, Franklin Lakes, NJ, USA). Briefly, transfected cells (5 × 104 cells/well) suspended in 200 μl serum-free medium were added to the top chamber. Then the lower chamber was added with 500 μl normal medium containing 10% FBS and incubated at 37 °C for 24 h. Cells moved to the lower chamber were fixed with 4% paraformaldehyde for 30 min and stained with 1% crystal violet for 30 min. Then the migrative and invasive abilities were quantitated by counting the cell numbers in five different areas under a light microscope (Olympus, Tokyo, Japan).
Real-time quantitative reverse transcription polymerase chain reaction (qRT-PCR)
Total RNA was extracted from tissues and cells using TRIzol reagent (Invitrogen). Then the obtained RNA (1 μg) was reversely transcribed into cDNA with a Transcription cDNA Synthesis Kit (Thermo Fisher Scientific, Waltham, MA, USA). Subsequently, the qPCR was performed with SYBR Premix Ex Taq (Takara Bio, Shiga, Japan) on an ABI 7500 fast real-time PCR system (Applied Biosystems, Foster, CA, USA). GAPDH was used as an endogenous control.
Target prediction
The target miRNA of DLEU1 and the target gene of miR-671-5p were predicted using bioinformatics online softwares starBase (http://starbase.sysu.edu.cn/index.php) and TargetScan (http://www.targetscan.org/vert_71/).
Luciferase reporter assay
To test the interaction between the DLEU1 and miR-671-5p, luciferase reporter assays were performed using the Dual-luciferase Reporter Assay System (Promega, Madison, WI, USA). The pGL3-reporter luciferase vector containing the sequence of wild-type or mutant type of DLEU1 was constructed and then co-transfected with miR-671-5p mimics or control mimics using Lipofectamine 2000 (Invitrogen). To assess the correlation between miR-671-5p and its target gene DDX5, the cells were co-transfected with pGL3 reporter luciferase vector containing the 3'-UTR sequence of wild-type or mutant type of DDX5 and miR-671-5p mimics or control mimics using Lipofectamine 2000 (Invitrogen). Cells were collected at 48 h after transfection and lysed for luciferase activity detection.
Statistical analysis
All data are presented as the mean ± SD calculated from three independent experiments. Statistical analysis was performed using SPSS 18.0 software with a Student’s test or one-way analysis of variance (ANOVA). For all comparisons, differences were considered significant when p-values less than .05.
Results
DLEU1 was highly expressed in osteosarcoma tissue specimens and cells
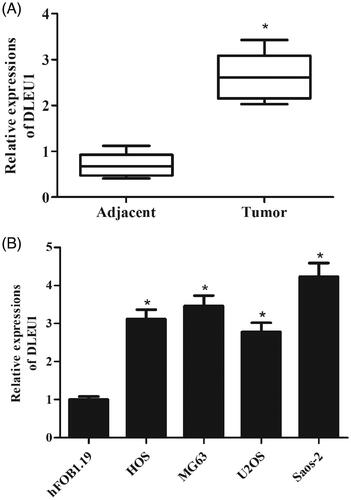
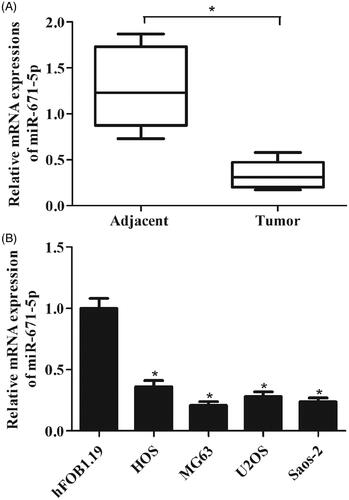
The qRT-PCR analysis was performed to examine the expression levels of DLEU1 in 50 paired osteosarcoma tissues and adjacent non-tumour tissues. The results showed that DLEU1 was frequently up-regulated in osteosarcoma tissues as compared to the adjacent non-tumour tissues (). In addition, the expression level of DLEU1 in hFOB1.19 cells was significantly lower than that in osteosarcoma cell lines HOS, MG63, U2OS, and Saos-2 cells ().
Figure 2. Expression levels of DLEU1 in specimens and cells. The expression levels of DLEU1 were examined using qRT-PCR analysis. (A) Expression levels of DLEU1 in 50 paired osteosarcoma tissues and adjacent non-tumour tissues. (B) Expression levels of DLEU1 in hFOB1.19 cells and osteosarcoma cell lines (HOS, MG63, U2OS, and Saos-2 cells). *p < .05.
Downregulation of DLEU1 inhibited the proliferation, migration and invasion of osteosarcoma cells
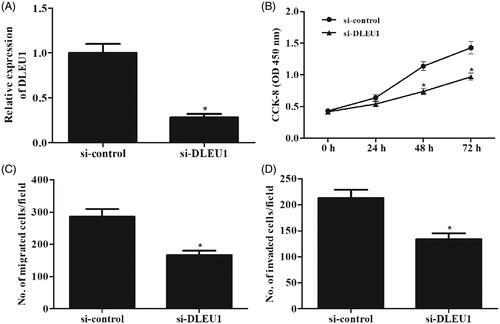
To investigate the role of DLEU1 in osteosarcoma, Saos-2 cells were transfected with si-DLEU1 or si-control. After 48 h post-transfection, expression levels of DLEU1 in Saos-2 cells were measured using qRT-PCR analysis. As shown in , DLEU1 expression was markedly decreased in Saos-2 cells transfected with si-DLEU1. Then CCK-8 assay was performed to determine cell proliferation. Downregulation of DLEU1 significantly repressed the proliferation of Saos-2 cells (). Transwell assay proved that cell migration and invasion were obviously decreased in si-DLEU1-transfected Saos-2 cells ().
Figure 3. Effects of DLEU1 on Saos-2 cells proliferation, migration and invasion. (A) Saos-2 cells were transfected with si-DLEU1 or si-control. The expression levels of DLEU1 in Saos-2 cells were measured using qRT-PCR analysis after 48 h post-transfection. (B) CCK-8 assay was performed to determine cell proliferation. (C, D) Transwell assay was carried out to detect cell migration and invasion. *p < .05.
DLEU1 targeted miR-671-5p in osteosarcoma cells
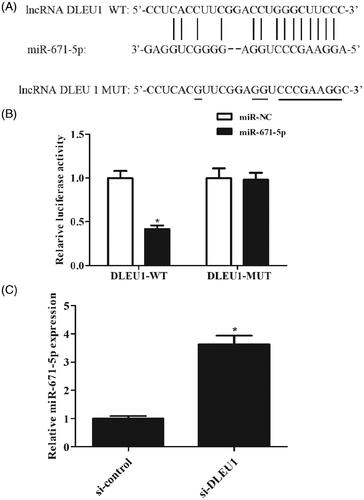
According to the online software, miR-671-5p might be a putative combining target miRNA of DLEU1 (). Dual-luciferase reporter assay was performed to explore the relation between DLEU1 and miR-671-5p. The results showed that co-transfection with pMIR-DLEU1-WT and miR-671-5p mimics greatly reduced the luciferase activity when compared with other groups (). In addition, downregulation of DLEU1 caused a significant increase in miR-671-5p expression in Saos-2 cells (). Therefore, we proposed DLEU1 might exert its function through regulating miR-671-5p in Saos-2 cells.
Figure 4. DLEU1 targeted miR-671-5p in Saos-2 cells. (A) MiR-671-5p might be a putative combining target miRNA of DLEU1 according to the prediction. (B) Dual-luciferase reporter assay was performed to confirm the relation between DLEU1 and miR-671-5p. (C) Effect of DLEU1 downregulation on miR-671-5p expression in Saos-2 cells. *p < .05.
miR-671-5p was lowly expressed in osteosarcoma tissue specimens and cells
We detected miR-671-5p expression in osteosarcoma tissues by qRT-PCR assay. The results indicated that miR-671-5p expression was markedly decreased in osteosarcoma tissues compared with the adjacent non-tumour tissues (). Similarly, we also found that miR-671-5p expression was significantly reduced in osteosarcoma cell lines ().
Figure 5. miR-671-5p was lowly expressed in osteosarcoma tissue specimens and cells. (A) The qRT-PCR analysis was performed to detect miR-671-5p expression in osteosarcoma tissues and the adjacent non-tumour tissues. (B) Expression levels of miR-671-5p in hFOB1.19 cells and osteosarcoma cell lines (HOS, MG63, U2OS, and Saos-2 cells). *p < .05.
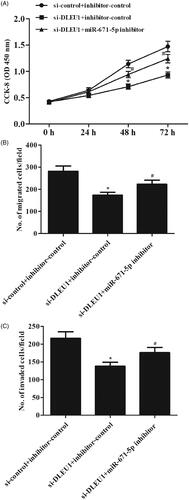
miR-671-5p inhibitor reversed the effects of si-DLEU1 on osteosarcoma cell proliferation and migration
Subsequently, we asked whether miR-671-5p mediated DLEU1’s effect on Saos-2 cell proliferation and invasion. The results showed that the inhibitory effects of DLEU1 down-regulation on cell proliferation, migration and invasion were rescued by anti-miR-671-5p treatment ().
Figure 6. miR-671-5p inhibitor reversed the effects of si-DLEU1 on osteosarcoma cell proliferation and migration. Saos-2 cells were transfected with si-DLEU1 (or si-control) and miR-671-5p inhibitors or controls. (A) Cell proliferation was detected using CCK-8 assay. (B, C) Cell migration and invasion abilities were detected with Transwell assay. *p < .05 vs. si-control + inhibitor-control; #p < .05 vs. si-DLEU1 + inhibitor-control.
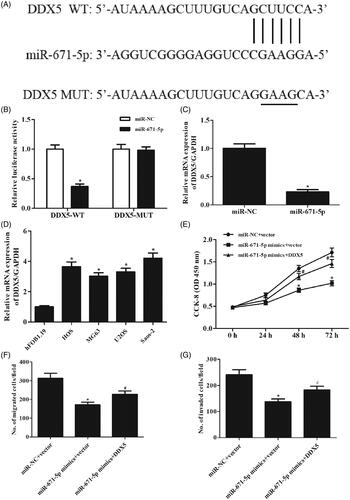
MiR-671-5p inhibited osteosarcoma cell proliferation, migration and invasion by downregulating the expression of DDX5
The online software indicated that DDX5 might be a target gene of miR-671-5p (). Results of dual-luciferase reporter assay demonstrated that miR-671-5p directly bound to the 3'-UTR of DDX5 (). Overexpression of miR-671-5p suppressed the expression of DDX5 in Saos-2 cells (). In addition, we examined the expression level of DDX5 in osteosarcoma cell lines and found that DDX5 expression was significantly increased in osteosarcoma cell lines, compared with the hFOB1.19 cell line (). Furthermore, we observed that restoration of DDX5 rescued miR-671-5p mimics effects on Saos-2 cell proliferation, migration and invasion (). These findings suggested that miR-671-5p exerted its roles by targeting DDX5.
Figure 7. MiR-671-5p inhibited osteosarcoma cell proliferation, migration and invasion by downregulating the expression of DDX5. (A) The binding sites between DDX5 and miR-671-5p predicted by bioinformatics website. (B) The binding relationship of DDX5 to miR-671-5p validated by dual-luciferase reporter gene assay. *p < .05 vs. miR-NC. (C) Effect of miR-671-5p mimics on DDX5 expression in Saos-2 cells. *p < .05 vs. miR-NC. (D) Relative expression of DDX5 in osteosarcoma cell lines. *p < .05 vs. Hfob1.19. Saos-2 cells were co-transfected with miR-671-5p mimics and DDX5 expression vector for different hours. (E) Cell proliferation was detected using CCK-8 assay. (F, G) Cell migration and invasion abilities were detected with Transwell assay. *p < .05 vs. miR-NC + vector; #p < .05 vs. miR-671-5p mimics + vector.
Discussion
Previous studies have revealed that many lncRNAs are involved in the pathogenesis of osteosarcoma. DLEU1 is a kind of lncRNA that has been found to be implicated in several cancers. In the present study, we investigated the role of DLEU1 in osteosarcoma. The results showed that DLEU1 is highly expressed in osteosarcoma tissue specimens and cells. Downregulation of DLEU1 inhibited the proliferation, migration and invasion of osteosarcoma cells. It has been documented that DLEU1 functions as an oncogene by regulating target miRNA. For instance, DLEU1 was found to be upregulated in bladder cancer tissues [Citation6]. Patients with high DLEU1 expression exhibit a shorter survival time. Further mechanistic analysis and functional assays validate that DLEU1 induces cell proliferation, invasion, and cisplatin resistance of bladder cancer cells through sponging miR-99b. Gao et al. [Citation14] reported that DLEU1 expression level is upregulated in pancreatic ductal adenocarcinoma (PDAC) tissues compared with adjacent normal tissue. The aberrant expression of DLEU1 indicates poor prognosis of PDAC patients. Loss-of-function experiments demonstrate that knockdown of DLEU1 inhibits the PDAC cells proliferation, migration, and invasion in vitro and suppresses the tumour growth in vivo. Further investigations indicate that DLEU1 aggravates PDAC carcinogenesis by sponging miR-381 [Citation14]. Wang et al. [Citation15] demonstrated that DLEU1 contributes to the tumourigenesis and development of ovarian carcinoma by interacting with miR-490-3p. Our results showed that miR-671-5p might be a putative combining target miRNA of DLEU1. Downregulation of DLEU1 caused a significant increase in miR-671-5p expression in Saos-2 cells. And, silenced of DLEU1 decreased cell proliferation, migration/invasion, but co-transfection with miR-671-5p inhibitor rescued these effects. The results indicated that DLEU1 might exert its role via sponging miR-671-5p.
MiR-671-5p has been found to be a tumour suppressor in several types of cancers [Citation16,Citation17]. Downregulation of miR-671-5p expression has been found in breast cancer tissues compared with their adjacent tissues [Citation18]. Overexpression of miR-671-5p inhibits breast cancer cell proliferation and invasion, as well results in a shift from epithelial-to-mesenchymal transition (EMT) to mesenchymal-to-epithelial transition (MET) phenotypes. In addition, the low expression levels of miR-671-5p are observed in gastric cancer cells [Citation19]. Overexpression of miR-671-5p inhibits cell proliferation and promotes cell apoptosis in gastric cancer cells. Our results demonstrated that miR-671-5p expression was markedly decreased in osteosarcoma tissues compared with the adjacent non-tumour tissues. MiR-671-5p overexpression inhibited cell proliferation, migration and invasion of Saos-2 cells. These findings suggested that miR-671-5p might be involved in osteosarcoma, which was attributed to the aberrant expression of DLEU1.
DDX5 is an ATP-dependent RNA helicase belonged to the DEAD-box protein family [Citation20]. Previous studies have suggested an important role for DDX5 in regulating transcription in conjunction with multiple, sequence-specific transcription factors [Citation21]. DDX5 is implicated in cancer development and progression by regulating cell proliferation, migration, cytoskeletal reorganization, and EMT [Citation22–26]. DDX5 has been found to be involved in the oncogenic processes of osteosarcoma. Chen et al. [Citation27] proved that the expressions of DDX5 are significantly higher in osteosarcoma patients tissues and osteosarcoma cells. Overexpression of DDX5 is associated with clinicopathological features and poor prognosis of osteosarcoma patients. Our results showed that DDX5 is a target gene of miR-671-5p in osteosarcoma cells. MiR-671-5p overexpression resulted in significant decrease in DDX5 expression, and restoration of DDX5 rescued miR-671-5p mimics effects on Saos-2 cell proliferation, migration and invasion. These data implying that miR-671-5p executed its tumour suppressive activity by targeting DDX5.
Our data defined a role for DLEU1 as an oncogene in osteosarcoma for the first time. DLEU1 acted its roles by directly sponging miR-671-5p and regulating the expression of DDX5. Collectively, DLEU1 might serve as a novel therapeutic target in the management of osteosarcoma.
Disclosure statement
The authors declare that there is no conflict of interest regarding the publication of this paper.
References
- Meyers PA, Gorlick R. Osteosarcoma. Pediatr Clin North Am. 1997;44:973–989.
- Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641.
- Li Z, Yu X, Shen J. Long non-coding RNAs: emerging players in osteosarcoma. Tumor Biol. 2016;37:2811–2816.
- Chen R, Wang G, Zheng Y, et al. Long non-coding RNAs in osteosarcoma. Oncotarget. 2017;8:20462–20475.
- Li JP, Liu LH, Li J, et al. Microarray expression profile of long noncoding RNAs in human osteosarcoma. Biochem Biophys Res Commun. 2013;433:200–206.
- Li Y, Shi B, Dong F, et al. Long non-coding RNA DLEU1 promotes cell proliferation, invasion, and confers cisplatin resistance in bladder cancer by regulating the miR-99b/HS3ST3B1 axis. Front Genet. 2019;10:280–288.
- Liu C, Tian X, Zhang J, et al. Long non-coding RNA DLEU1 promotes proliferation and invasion by interacting with miR-381 and enhancing HOXA13 expression in cervical cancer. Front Genet. 2018;9:629.
- Nishiyama K, Maruyama R, Niinuma T, et al. Screening for long noncoding RNAs associated with oral squamous cell carcinoma reveals the potentially oncogenic actions of DLEU1. Cell Death Dis. 2018;9:826.
- Zhang S, Guan Y, Liu X, et al. Long non-coding RNA DLEU1 exerts an oncogenic function in non-small cell lung cancer. Biomed Pharmacother. 2019;109:985–990.
- Liu T, Han Z, Li H, et al. LncRNA DLEU1 contributes to colorectal cancer progression via activation of KPNA3. Mol Cancer. 2018;17:118.
- Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159.
- Wang K, Chang H. Molecular mechanisms of long noncoding RNAs. Molecul Cell. 2011;43:904–914.
- Fang Y, Fullwood MJ. Roles, functions, and mechanisms of long non-coding RNAs in cancer. Genom Proteom Bioinf. 2016;14:42–54.
- Gao S, Cai Y, Zhang H, et al. Long noncoding RNA DLEU1 aggravates pancreatic ductal adenocarcinoma carcinogenesis via the miR-381/CXCR4 axis. J Cell Physiol. 2019;234:6746–6757.
- Wang LL, Sun KX, Wu DD, et al. DLEU1 contributes to ovarian carcinoma tumourigenesis and development by interacting with miR-490-3p and altering CDK1 expression. J Cell Mol Med. 2017;21:3055–3065.
- Malgulwar PB, Pathak P, Singh M, et al. Downregulation of SMARCB1/INI1 expression in pediatric chordomas correlates with upregulation of miR-671-5p and miR-193a-5p expressions. Brain Tumor Pathol. 2017;34:155–159.
- Barbagallo D, Condorelli A, Ragusa M, et al. Dysregulated miR-671-5p/CDR1-AS/CDR1/VSNL1 axis is involved in glioblastoma multiforme. Oncotarget. 2016;7:4746–4759.
- Tan X, Fu Y, Chen L, et al. miR-671-5p inhibits epithelial-to-mesenchymal transition by downregulating FOXM1 expression in breast cancer. Oncotarget. 2016;7:293–307.
- Qiu T, Wang K, Li X, et al. miR-671-5p inhibits gastric cancer cell proliferation and promotes cell apoptosis by targeting URGCP. Exp Ther Med. 2018;16:4753–4758.
- Giraud G, Terrone S, Bourgeois CF. Functions of DEAD box RNA helicases DDX5 and DDX17 in chromatin organization and transcriptional regulation. BMB Rep. 2018;51:613–622.
- Fuller-Pace FV. The DEAD box proteins DDX5 (p68) and DDX17 (p72): multi-tasking transcriptional regulators. Biochim Biophys Acta. 2013;1829:756–763.
- Du C, Li DQ, Li N, et al. DDX5 promotes gastric cancer cell proliferation in vitro and in vivo through mTOR signaling pathway. Sci Rep. 2017;7:42876.
- Wang Z, Luo Z, Zhou L, et al. DDX5 promotes proliferation and tumorigenesis of non-small-cell lung cancer cells by activating beta-catenin signaling pathway. Cancer Sci. 2015;106:1303–1312.
- Ma Z, Feng J, Guo Y, et al. Knockdown of DDX5 inhibits the proliferation and tumorigenesis in esophageal cancer. Oncol Res. 2017;25:887–895.
- Mazurek A, Luo W, Krasnitz A, et al. DDX5 regulates DNA replication and is required for cell proliferation in a subset of breast cancer cells. Cancer Discov. 2012;2:812–825.
- Nyamao RM, Wu J, Yu L, et al. Roles of DDX5 in the tumorigenesis, proliferation, differentiation, metastasis and pathway regulation of human malignancies. Biochim Biophys Acta Rev Cancer. 2019;1871:85–98.
- Chen Y, Wang Q, Wang Q, et al. DEAD-Box helicase 5 interacts with transcription factor 12 and promotes the progression of osteosarcoma by stimulating cell cycle progression. Front Pharmacol. 2018;9:1558.