Abstract
Background
Non-small-cell lung carcinoma (NSCLC) is the most common type of lung cancer. Circular RNA ZFR (circZFR) is an identified circular RNA (circRNA) that is correlated with cancer progression. However, the role of circZFR in NSCLC remains unknown. In the present study, we aimed to investigate the function of circZFR in NSCLC and the underlying mechanism.
Methods
The expression patterns of circZFR were determined using qRT-PCR in NSCLC samples and cell lines. The subcellular distribution of circZFR in NSCLC cells was analyzed by fluorescence in situ hybridization (FISH). Cell proliferation was examined utilizing the CCK-8 assay. Cell migration and invasion were evaluated using the Transwell assay. We used the bioinformatics software TargetScan and miRanda to predict circRNA–miRNA and miRNA–mRNA interactions.
Results
Our results showed that circZFR expressions were significantly upregulated in NSCLC tissues and cell lines. Knockdown of circZFR significantly inhibited the cell proliferation, migration and invasion of NSCLC cells in vitro. Furthermore, we demonstrated that circZFR acted as a sponge to absorb microRNA-101-3p (miR-101-3p) and promoted cullin 4B (CUL4B) expression.
Conclusions
Collectively, our results demonstrated that circZFR exhibited a carcinogenic role by sponging miR-101-3p and regulating CUL4B expression in NSCLC. These findings provided evidence for understanding the role of circZFR in NSCLC tumourigenesis.
Introduction
Lung cancer is a malignant lung tumour that remains the leading cause of cancer-related mortality worldwide [Citation1]. Lung cancer is characterized by uncontrolled cell growth in lung tissues; moreover, this growth can spread into nearby tissues or other tissues by the process of metastasis. There are two main types of lung cancer, which are small-cell lung carcinoma (SCLC) and non-small-cell lung carcinoma (NSCLC). NSCLC is the most common type and accounts for about 85% of all lung cancers [Citation2]. There are different types of treatment options for patients with NSCLC, depending on the stage of cancer. The currently used treatments include surgery, radiation therapy, chemotherapy, targeted therapy, immunotherapy and combined therapy [Citation3,Citation4]. However, the treatments for the advanced/metastatic NSCLC should be developed. Therefore, better understanding of the pathogenesis of NSCLC may be beneficial for developing new therapeutic approaches.
Emerging pieces of evidence have demonstrated that carcinogens cause genetic mutations or dysregulations that induce the development of cancer [Citation5]. NSCLC has been found to be initiated by either the activation of oncogenes or the inactivation of tumour suppressor genes [Citation6]. Circular RNA (circRNA) is a type of non-coding RNA, which is characterized by the presence of a covalent bond linking the 3’ and 5’ ends [Citation7]. Previous studies proved that circRNAs are not simply a side product of splicing. More and more studies indicated that circRNAs participate in various cellular processes [Citation8]. CircRNAs act as microRNA (miRNA) sponges and RNA-binding protein sequestering agents as well as nuclear transcriptional regulators, thereby governing gene expression [Citation8]. A growing body of evidence showed that dysregulated expression of circRNAs plays critical roles in the development of several human diseases, including cancer [Citation9–11].
Circular RNA ZFR (circZFR) is an identified cancer-related circRNA through sponging miRNAs. Liu et al. [Citation12] reported that circZFR inhibits cell proliferation and induces apoptosis by sponging miR-130a/miR-107 and modulating PTEN in gastric cancer. Tan et al. [Citation13] proved that circZFR exerts carcinogenic role in hepatocellular carcinoma (HCC) progression through regulating miR-3619-5p/CTNNB1 axis and activating Wnt/β-catenin pathway. Besides, circZFR was found to regulate cell proliferation and invasion by sponging miR-1261 and facilitating C8orf4 expression in papillary thyroid cancer (PTC) [Citation14]. Nonetheless, the regulatory function of circZFR on NSCLC remains unknown. In the present study, we aimed to investigate the role of circZFR in NSCLC.
Materials and methods
Clinical tissues and cell culture
A total of 15 NSCLC tissues and corresponding adjacent normal lung tissues were collected from NSCLC patients who underwent surgeries at the Department of Thoracic Surgery, Huaihe Hospital of Henan University (Kaifeng, China), between January 2016 and February 2017. All participants were diagnosed as primary NSCLC based on the postoperative histopathological findings without any previous anticancer treatment. All tissue samples were snap-frozen in liquid nitrogen and stored at −80 °C until total RNA extraction. The informed consent and agreement for the samples’ usage were obtained from all enrolled patients. The study was permitted by the Medical Ethical Committee of the Huaihe Hospital of Henan University.
A normal human bronchial epithelial cell line (HBE), and five NSCLC cell lines A549, H460, H1703, PC9 and H1299 were obtained from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). HEK-293T cells were obtained from ATCC (Manassas, VA). Cells were cultured in RPMI-1640 medium (Hyclone, Logan, UT) supplemented with 10% heat-inactivated fetal bovine serum (FBS, Hyclone), 100 U/ml penicillin and 100 mg/ml streptomycin (Sigma-Aldrich, St. Louis, MO), and maintained in a humidified incubator with 5% CO2 at 37 °C.
Cell transfection
A549 and H1299 cells were cultured in 6-well plates and then transfected with a short interfering RNA targeting circZFR (si-circZFR)/control siRNA (si-NC; 100 nM), miR-101-3p inhibitor/control inhibitor, miR-101-3p mimics/negative control (NC), cullin 4B (CUL4B)-overexpressing plasmid/plasmid (Shanghai Genepharma Biotech Co., Ltd., Shanghai, China) using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. For si-circZFR, the sequence of was 5′-CAAATTTATGCCCAGCCGGCT-3′.
Cell proliferation assay
Cell proliferation was determined using the Cell Counting Kit-8 (CCK-8) assay kit (Beyotime, Shanghai, China). A549 and H1299 cells (5000 cells/well) were plated in 96-well plates and allowed to grow for 0, 24, 48, 72 or 96 h after transfection. Then 10 μl of CCK8 solution was added to each well and incubated for 3 h. The absorbance was assessed at a wavelength of 450 nm using a microplate reader (Bio-Tek, Winooski, VT).
Cell migration and invasion assays
Transwell assay was performed to evaluate cell migration and invasion using Transwell chambers. For the invasion assay, chambers were coated with Matrigel. A549 and H1299 cells were seeded into the upper chamber (2000 cells), while the lower chamber was filled with complete medium. After incubation for 24 h, the residual cells on the upper side of the filters were removed, and cells on the lower side of the filter were fixed with 4% paraformaldehyde for 30 min and then stained with crystal violet for 10 min. Four random images were captured and the cell numbers were counted.
qRT-PCR
Total RNAs were extracted from clinical tissues and cell lines using TRIzol reagent (Invitrogen). 2 μg RNA of each sample was reverse transcribed using SuperScript RT kit (Invitrogen, Carlsbad, CA, USA). qRT-PCR was performed on an Applied Biosystems 7300 Real-time PCR system (Applied Biosystems, CA) using SYBR Premix Ex Taq™ (Takara, Japan). GAPDH or U6 were used as internal references. The relative expressions of target genes were calculated using the 2−ΔΔCt method. The sequences of the primers used in this study are listed as follows: circZFR forward, 5’-AACCACCACAGATTCACTAT-3’, reverse, 5’-AACCACCACAGATTCACTAT-3’; miR-101-3p forward, 5’-ACGGGCGAGCTACAGTACTGTG-3’, reverse, 5’-CCAGTGCAGGGTCCGAGGTA-3’; CUL4B forward, 5’-GGGAAAGGAATGGTGAA-3’, reverse, 5’-TGCATAGAGCCGGTTAG-3’; GAPDH forward, 5’-ATAGCACAGCCTGGATAGCAACGTAC-3’, reverse, 5’-CACCTTCTACAATGAGCTGCGTGTG-3’; U6 forward, 5’-CGCTTCGGCAGCACATATAC-3’, reverse, 5’-TTCACGAATTTGCGTGTCAT-3’.
Western blot
Total cell lysates of A549 and H1299 cells were prepared by using radioimmunoprecipitation assay (RIPA) buffer (Solarbio, Beijing, China) according to the manufacturer’s protocols. Protein concentrations in the lysates were measured using a bicinchoninic acid assay (BCA) kit (Beyotime Biotechnology, Shanghai, China). Equal amounts of total protein were separated by 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), and subsequently transferred to polyvinylidene fluoride (PVDF) membranes (Millipore, Bedford, MA, USA). Then the membranes were incubated with specific primary antibodies targeting CUL4B (1:500, Abcam, Cambridge, MA) and GAPDH (1:1000, Abcam) overnight at 4 °C. After that, the membranes were probed with horseradish peroxidase-conjugated secondary antibody (1:3000, Abcam) at room temperature for 1 h. The signal on the membranes was visualized using ECL reagent (Thermo). The intensity was measured using Image J analysis software (National Institutes of Health, NIH, Bethesda, MD).
Target prediction
The miRNAs that interact with circZFR were predicted from circular RNA interactome and Targetscan. The target genes of miR-101-3p were predicted by Targetscan and mirTarbase.
Luciferase reporter assay
The fragments of circZFR and CUL4B containing the miR-101-3p binding sites were synthesized by Sangon Biotech (Shanghai, China), which were named WT-circZFR or WT-CUL4B. The corresponding mutated fragments, MUT-circZFR or MUT-CUL4B were also synthesized. These wide or mutated type fragments were sub-cloned into the reporter vector psiCHECK-2 (Promega, Madison, WI, USA). HEK-293T cells (1 × 105 cells/well) were seeded into 24-well plates and then co-transfected with miR-101-3p mimics/control mimics and WT-circZFR/MUT-circZFR or WT-CUL4B/MUT-CUL4B using Lipofectamine 2000 (Invitrogen). After 24 h post-transfection, luciferase activity was measured using the dual-luciferase reporter assay system (Promega).
Fluorescence in situ hybridization (FISH)
Biotin-labelled circZFR probe was designed and synthesized by RiboBio (Guangzhou, China). The signals of the biotin-labelled probes were detected using Cy5-conjugated streptavidin (Life Technologies, Waltham, MA). Cell nuclei were counterstained with 4,6-diamidino-2-phenylindole (DAPI). Finally, the images were obtained on a Zeiss LSM 700 confocal microscope (Carl Zeiss, Oberkochen, Germany).
Statistical analysis
Data are presented as the means ± standard deviation (SD). The SPSS software (version 20.0, SPSS, Inc., Chicago) was applied for the data analysis. The Student’s t-test or one-way analysis of variance (ANOVA) was used for the statistical analysis, and p < .05 was considered statistically significant.
Results
Knockdown of circZFR inhibited the proliferation, migration and invasion of NSCLC cells
The expression levels of circZFR in clinical tissue specimens were measured using qRT-PCR. As shown in , circZFR expression was significantly elevated in NSCLC tissues compared with adjacent normal lung tissues. Then we examined the expression levels of circZFR in NSCLC cell lines. CircZFR expression was markedly upregulated in NSCLC cell lines, especially in A549 and H1299 cells (. Fluorescence in situ hybridization (FISH) assay revealed that circZFR predominately localized in the cytoplasm (Supplementary Figure S1).
Figure 1. Knockdown of circZFR inhibited the proliferation, migration and invasion of NSCLC cells. (A) Expression levels of circZFR in NSCLC tissues and adjacent normal tissues. *p < .05 vs. adjacent normal lung tissues. (B) Expression levels of circZFR in HBE cell line and NSCLC cell lines. *p < .05 vs. HBE cell line. (C, D) Transfection efficiency was confirmed using qRT-PCR after transfection with si-circZFR or si-NC in A549 and H1299 cells. (E, F) Cell proliferation of A549 and H1299 cells was measured using CCK-8 assay. (G, H) Cell migration and invasion of A549 and H1299 cells were assessed using the Transwell assay. *p < .05 vs. cells transfected with si-NC.
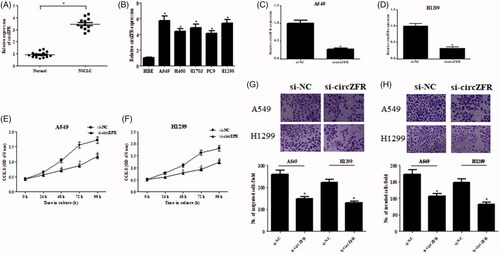
To further identify the role of circZFR in NSCLC, circZFR was knocked down in A549 and H1299 cells through transfection with si-circZFR. The knockdown efficiency was confirmed using qRT-PCR (. Next, we examined the effects of circZFR knockdown on cell proliferation, migration and invasion in A549 and H1299 cells. As illustrated in , knockdown of circZFR significantly inhibited the cell proliferation, migration and invasion of NSCLC cells.
CircZFR might regulate CUL4B expression by directly targeting miR-101-3p
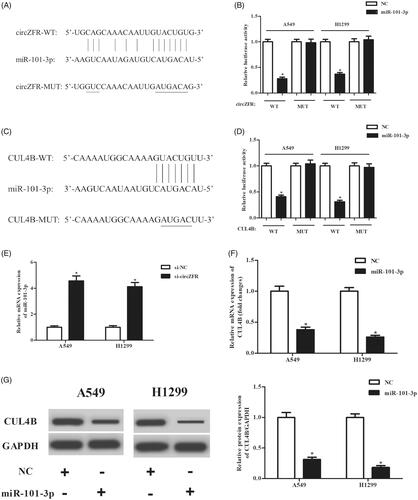
According to the predicted results from online software, circZFR might act as a sponge of miR-101-3p (. Furthermore, CUL4B might be a target gene of miR-101-3p (. To validate these predictions, luciferase reporter assays were performed. The results in indicated that overexpression of miR-101-3p significantly repressed the luciferase activity when co-transfected with WT-circZFR or WT-CUL4B. In addition, the expression levels of miR-101-3p were markedly increased after transfection with si-circZFR (. Besides, overexpression of miR-101-3p significantly inhibited the expression of CUL4B (.
Figure 2. CircZFR regulated CUL4B expression by directly targeting miR-101-3p. (A) The putative sequences of miR-101-3p and circZFR. (C) The putative sequences of miR-101-3p and the 3’-UTR of CUL4B mRNA. (B, D) Luciferase reporter assay was performed to validate these predictions. *p < .05 vs. NC group. (E) Expression levels of miR-101-3p after transfection with si-circZFR or si-NC. *p < .05 vs. si-NC group. (F) Expression levels of CUL4B after transfection with miR-101-3p mimics or control mimics. *p < .05 vs. negative control (NC) group.
The effects of circZFR knockdown on NSCLC cells were abolished by miR-101-3p inhibitor
Next, we detected the expression levels of miR-101-3p in human NSCLC cell lines. As shown in , miR-101-3p was downregulated in NSCLC cell lines. Furthermore, the Spearman rank correlation assessment indicated an inverse correlation between circZFR and miR-101-3p in NSCLC tissues ().
Figure 3. MiR-101-3p inhibitor prevented the effects of circZFR knockdown on NSCLC cells. (A) Expression levels of miR-101-3p in human NSCLC cell lines. *p < .05. (B) Transfection efficiency was confirmed after transfection with miR-101-3p inhibitor or control inhibitor in si-circZFR transfected A549 and H1299 cells. *p < .05 vs. si-NC + inhibitor-control; #p < .05 vs. si-circZFR + inhibitor-control. (C, D) Cell proliferation of A549 and H1299 cells was measured using CCK-8 assay. (E, F) Cell migration and invasion of A549 and H1299 cells was assessed using the Transwell assay. *p < .05 vs. si-NC + inhibitor-control; #p < .05 vs. si-circZFR + inhibitor-control.
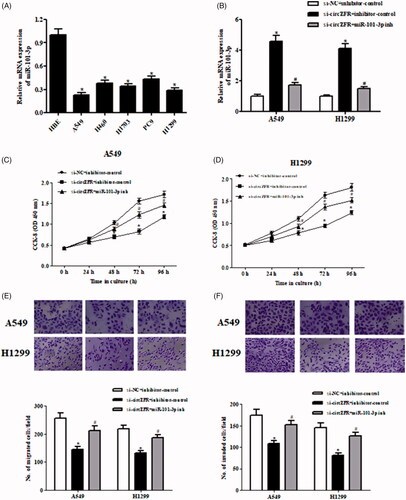
To further investigate the role of miR-101-3p in the effects of circZFR, miR-101-3p inhibitor was transfected into A549 and H1299 cells. After transfection with miR-101-3p inhibitor, the expression levels of miR-101-3p were dramatically decreased (. Downregulation of miR-101-3p prevented the si-circZFR-caused inhibitory effects on cell proliferation, migration and invasion in A549 and H1299 cells ().
CUL4B overexpression abolished the effects of miR-101-3p on NSCLC cells
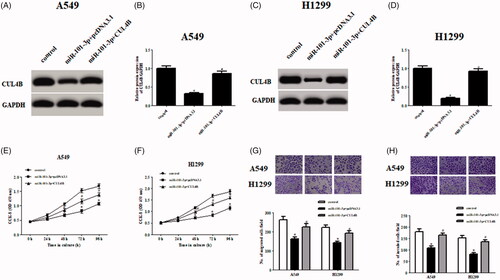
To investigate whether miR-101-3p exerts roles by targeting CUL4B in NSCLC cells, we restored CUL4B expression by transfection with CUL4B-overexpressing plasmid in A549 and H1299 cells. Overexpression of CUL4B significantly increased the expression of CUL4B in miR-101-3p transfected A549 and H1299 cells (). Further investigations revealed that the decreased cell proliferation, migration and invasion in miR-101-3p-overexpressed cells were significantly prevented by overexpression of CUL4B ().
Figure 4. CUL4B overexpression abolished the effects of miR-101-3p on NSCLC cells. (A–D) Protein expression levels of CUL4B after transfection with CUL4B-overexpressing plasmid in miR-101-3p transfected A549 and H1299 cells. (E–H) Cell proliferation, migration and invasion of A549 and H1299 cells were measured using CCK-8 assay and Transwell assay, respectively. *p < .05 vs. control; #p < .05 vs. miR-101-3p + pcDNA3.1.
Discussion
CircRNAs are a wide category of ncRNAs that exhibit broad biological activities and widely involved in numerous physiological and pathological processes. Currently, circRNAs have become a hotspot in the field of cancer biology [Citation15,Citation16]. Some findings indicated that dysregulated circRNAs had been verified to be associated with NSCLC progression. For instance, expression levels of circRNA_100876 are significantly elevated in NSCLC tissues as compared to the adjacent non-tumourous tissues. The up-regulated expression of circRNA_100876 is associated with lymph node metastasis and tumour staging in patients with NSCLC. Furthermore, the overall survival time of NSCLC patients with higher circRNA_100876 expression is significantly shorter than those patients with lower circRNA_100876 expression. These findings indicated that circRNA_100876 is correlated with the carcinogenesis of NSCLC [Citation17]. Liu et al. [Citation18] reported that circ_0001649 expression is decreased in NSCLC tissues and cell lines. The downregulation is correlated with advanced TNM stage, positive lymph node metastasis and unfavourable prognosis. Additionally, circ_0001649 inhibits cell growth and metastasis both in vitro and in vivo. This study suggested that circ_0001649 acts as a prognostic biomarker and tumour suppressor in NSCLC progression.
CircZFR is a circRNA that has been identified to be implicated in the development of several cancers. CircZFR is low-expressed in gastric cancer tissues and cell lines. Overexpression of circZFR inhibits cell proliferation, arrests cell cycle and promotes cell apoptosis by sponging miR-107/miR-130a in gastric cancer cells. In addition, miR-107/miR-130a regulates cell proliferation and cell apoptosis through targeting PTEN, which is a tumour suppressor. In vivo study proved that circZFR inhibits GC tumour growth and affects the expressions of p53 and PTEN [Citation12]. CircZFR is significantly upregulated in PTC tissues compared to adjacent normal tissues. The circZFR expression level is negatively correlated with clinical severity. Knockdown of circZFR dramatically inhibits the cell proliferation, migration and invasion of PTC cells. The following experiments demonstrated that circZFR exerts its oncogenic roles by regulating miR-1261/C8orf4 axis in PTC [Citation14]. Besides, circZFR was found to be closely related to HCC progression. Moreover, high expression of circZFR is associated with the poor prognosis of patients with HCC. Further loss-of-function assay demonstrated that knockdown of circZFR in HCC cells significantly suppresses cell proliferation and epithelial-mesenchymal transition (EMT) process. in addition, circZFR executes its carcinogenic role in HCC through regulating the CTNNB1 expression via sponging miR-3619-5p [Citation13]. In the present study, we found that circZFR expression was markedly upregulated in NSCLC tissues and cell lines. Knockdown of circZFR significantly inhibited the proliferation, migration and invasion of NSCLC cells.
It has been found that circRNAs can bind to miRNA and act as RNA sponge, thereby regulate downstream gene expression, thus contributing to the development of many cancers [Citation15]. As discussed above, circZFR executes its carcinogenic and tumour suppressive roles by sponging miRNAs. MiR-101-3p, a member of the miR-101 family, has been observed to be involved in the development of various cancers, such as breast cancer, glioblastoma, HCC and NSCLC [Citation19–22]. A recent study revealed that miR-101-3p has tumour suppressor effects in patients with NSCLC. The mir-101-3p expression is down-regulated in NSCLC and its expression level is a crucial risk factor for overall survival (OS) and disease-free survival (DFS) rate in patients with NSCLC. Besides, the miR-101-3p expression is associated with tumour diameter, lymph node metastasis and tumour-node-metastasis stage, indicating that miR-101-3p may serve as a biomarker for patients with NSCLC [Citation22]. Moreover, overexpression of miR-101-3p significantly inhibits cell proliferation, migration and invasion of NSCLC cells. The in vivo study showed that miR-101-3p results in the inhibition on the growth and metastasis of NSCLC cells [Citation23]. In the present study, the results showed that miR-101-3p expression was obviously downregulated in both NSCLC tissues and cell lines. Furthermore, we found that circZFR executes its carcinogenic role in NSCLC by sponging miR-101-3p.
CUL4B, a scaffold protein, is aberrantly expressed in diverse types of cancers, including NSCLC. Mi et al. [Citation24] demonstrated that CUL4B is upregulated in NSCLC tissues. It is critically required for cell proliferation and migration in vitro and for xenograft tumour formation in vivo. Wang et al. also reported that CUL4B is highly expressed in NSCLC cell lines. Silencing CUL4B inhibits proliferation, migration/invasion and EMT progress through suppressing the Wnt/β-catenin signalling pathway in NSCLC cells [Citation25]. In the current study, we showed that CUL4B was a target gene of miR-101-3p. Further investigations revealed that miR-101-3p overexpression-caused decreases in cell proliferation, migration and invasion were significantly prevented by overexpression of CUL4B. The results indicated that miR-101-3p exerted its tumour suppressor effects by targeting CUL4B in NSCLC.
Conclusions
Collectively, our study showed that circZFR was upregulated in NSCLC tissues and cell lines. Knockdown of circZFR in NSCLC cells significantly inhibited cell proliferation, migration and invasion. Furthermore, we demonstrated that circZFR executed a carcinogenic role by regulating miR-101-3p/CUL4B axis.
Disclosure statement
The authors declare that they have no competing interests.
Additional information
Funding
References
- Duruisseaux M, Esteller M. Lung cancer epigenetics: from knowledge to applications. Semin Cancer Biol. 2018;51:116–128.
- Tsim S, O'Dowd CA, Milroy R, et al. Staging of non-small cell lung cancer (nsclc): a review. Respir Med. 2010;104:1767–1774.
- Watanabe SI, Nakagawa K, Suzuki K. Neoadjuvant and adjuvant therapy for stage III non-small cell lung cancer. Jpn J Clin Oncol. 2017;47:1112–1118.
- PDQ Adult Treatment Editorial Board. Non-small cell lung cancer treatment (PDQ®): patient version. Bethesda (MD): National Cancer Institute; 2002; (PDQ cancer information summaries).
- Kutkowska J, Porębska I, Rapak A. Non-small cell lung cancer - mutations, targeted and combination therapy. Postepy Hig Med Dosw. 2017;71:431–445.
- Hirsch FR, Suda K, Wiens J, et al. New and emerging targeted treatments in advanced non-small-cell lung cancer. Lancet. 2016;388:1012–1024.
- Ebbesen KK, Kjems J, Hansen TB. Circular RNAs: identification, biogenesis and function. Biochim Biophys Acta. 2016;1859:163–168.
- Liu L, Wang J, Khanabdali R, et al. Circular RNAs: isolation, characterization and their potential role in diseases. RNA Biol. 2017;14:1715–1721.
- Haque S, Harries LW. Circular RNAs (circRNAs) in health and disease. Genes. 2017;8:1–17.
- Greene J, Baird AM, Brady L. Circular RNAs: biogenesis, function and role in human diseases. Front Mol Biosci. 2017;4:1–11.
- Patop IL, Kadener S. CircRNAs in cancer. Curr Opin Genet Dev. 2018;48:121–127.
- Liu T, Liu S, Xu Y, et al. Circular RNA-ZFR inhibited cell proliferation and promoted apoptosis in gastric cancer by sponging mir-130a/mir-107 and modulating PTEN. Cancer Res Treat. 2018;50:1396–1417.
- Tan A, Li Q, Chen L. CircZFR promotes hepatocellular carcinoma progression through regulating mir-3619-5p/ctnnb1 axis and activating Wnt/β-catenin pathway. Arch Biochem Biophys. 2019;661:196–202.
- Wei H, Pan L, Tao D, et al. Circular RNA circZFR contributes to papillary thyroid cancer cell proliferation and invasion by sponging mir-1261 and facilitating c8orf4 expression. Biochem Biophys Res Commun. 2018;503:56–61.
- Zhu LP, He YJ, Hou JC, et al. The role of circRNAs in cancers. Biosci Rep. 2017;37:1–21.
- Qian L, Yu S, Chen Z, et al. The emerging role of circRNAs and their clinical significance in human cancers. Biochim Biophys Acta Rev Cancer. 2018;1870:247–260.
- Yao JT, Zhao SH, Liu QP, et al. Over-expression of circRNA_100876 in non-small cell lung cancer and its prognostic value. Pathol Res Pract. 2017;213:453–456.
- Liu T, Song Z, Gai Y. Circular rna circ_0001649 acts as a prognostic biomarker and inhibits nsclc progression via sponging mir-331-3p and mir-338-5p. Biochem Biophys Res Commun. 2018;503:1503–1509.
- Liu P, Ye F, Xie X, et al. Mir-101-3p is a key regulator of tumor metabolism in triple negative breast cancer targeting ampk. Oncotarget. 2016;7:35188–35198.
- Li L, Shao MY, Zou SC, et al. Mir-101-3p inhibits EMT to attenuate proliferation and metastasis in glioblastoma by targeting TRIM44. J Neurooncol. 2019;141:19–30.
- Moshiri F, Salvi A, Gramantieri L, et al. Circulating mir-106b-3p, mir-101-3p and mir-1246 as diagnostic biomarkers of hepatocellular carcinoma. Oncotarget. 2018;9:15350–15364.
- Lu HM, Yi WW, Ma YS, et al. Prognostic implications of decreased microRNA-101-3p expression in patients with non-small cell lung cancer. Oncol Lett. 2018;16:7048–7056.
- Zhang X, He X, Liu Y, et al. Mir-101-3p inhibits the growth and metastasis of non-small cell lung cancer through blocking pi3k/akt signal pathway by targeting malat-1. Biomed Pharmacother. 2017;93:1065–1073.
- Mi J, Zou Y, Lin X, et al. Dysregulation of the mir-194-cul4b negative feedback loop drives tumorigenesis in non-small-cell lung carcinoma. Mol Oncol. 2017;11:305–319.
- Wang X, Chen Z. Knockdown of cul4b suppresses the proliferation and invasion in non-small cell lung cancer cells. Oncol Res. 2016;24:271–277.
