Abstract
This study aimed to investigate the effect and potential modulation mechanism of lnc-SLC4A1-1 on breast cancer (BC) carcinogenesis. The expression of lnc-SLC4A1-1 in tissue and serum samples from BC patients, as well as BC cell lines, was detected by real-time quantitative reverse transcription-polymerase chain reactions (qRT-PCRs). Next, the expression of lnc-SLC4A1-1 was silenced or upregulated in BC cells, then cell proliferation, apoptosis, migration and invasion were detected using MTT, flow cytometry analysis and Transwell assay. Meanwhile, the expression of apoptosis-related proteins and epithelial-mesenchymal transition-related proteins were detected by western blotting. Furthermore, potential mechanism of lnc-SLC4A1-1 was explored by chromatin immunoprecipitation and RNA immunoprecipitation assays. CXCL8 was overexpressed to evaluate the relationship between lnc-SLC4A1-1 and CXCL8. Lnc-SLC4A1-1 was significantly up-regulated in BC tissue, serum samples and cell lines. In BC cells, lnc-SLC4A1-1 knockdown promoted cell apoptosis and suppressed cell proliferation, migration and invasion. Furthermore, lnc-SLC4A1-1 is transcriptionally activated by H3K27 acetylation, and lnc-SLC4A1-1 interacted with transcription factor (NF)-κB p65, thereby regulating CXCL8 expression. Meanwhile, CXCL8 overexpression partly reversed the effects of lnc-SLC4A1-1 knockdown on cell viability, apoptosis, migration and invasion in BC cells. Lnc-SLC4A1-1 could promote the development of BC by regulating NF-κB/CXCL8.
Lnc-SLC4A1-1 was overexpressed in BC tissues, blood and cell lines.
Lnc-SLC4A1-1 was transcriptionally activated by H3K27 acetylation.
Lnc-SLC4A1-1 interacted with NF-κB to promote CXCL8 expression.
Lnc-SLC4A1-1 could promote the development of BC.
Highlights
Introduction
Breast cancer (BC) is a common malignant tumor originating in the epithelial tissues of the breast gland among women and is counted as one of the top ten common cancers worldwide [Citation1]. Evidence from BC epidemiology estimates ∼1.7 million new diagnosed cases with high mortality in 2016 [Citation2]. Nowadays, the conventional and effective therapeutic methods for BC include targeted pharmacological treatments, sophisticated surgical resection and advanced chemotherapy [Citation3]. Despite the intensive improvements in diagnosis and treatments, poor survival and unsatisfactory prognosis remain an issue due to delayed diagnosis, high rates of recurrence and metastasis, and adverse drug effects [Citation4]. Therefore, it is important to further reveal novel diagnostic and therapeutic methods, as well as investigate the underlying molecular mechanism of BC.
Encouragingly, overwhelming evidence has demonstrated that the regulatory roles of noncoding RNAs such as circular RNAs (circRNAs), long non-coding RNAs (lncRNAs) and microRNAs (miRNAs) are predominately correlated with the development and progression of a wide variety of cancers [Citation5]. LncRNAs, one class of noncoding RNA with the length > 200 nucleotides, have been concerned by plenty of researches, which have revealed several possible functions, including miRNA sponges, regulating gene transcription and splicing, and forming RNA-protein complexes [Citation6]. It has been widely acknowledged that lncRNAs may participate in disease progression by regulating various key cell biological process such as cell proliferation, differentiation, apoptosis, migration and invasion [Citation7]. Remarkably, the prevalence of high-throughput sequencing technologies makes diverse bioinformatics analyses available, and study has revealed dysregulated expression of lncRNAs in many cancers [Citation8]. An integrated transcriptome analysis has identified several BC-specific lncRNAs, and the following in vitro experiments have demonstrated that lncRNAs HIF1A-AS2 and AK124454 are involved in the development of triple-negative BC through promoting cell proliferation and invasion [Citation9]. Noteworthily, a recent study has revealed that lnc-SLC4A1-1 as enhancer RNA epigenetically alters trophoblast function in unexplained recurrent pregnancy loss via regulating chemotactic factor CXCL8 and nuclear factor (NF)-kB pathway [Citation10]. However, the function and mechanism of lnc-SLC4A1-1 in BC is still unclear.
In the present study, the expression level of lnc-SLC4A1-1 has identified in BC tumor and blood samples as well as BC cells. Then the effects of lnc-SLC4A1-1 on BC cell proliferation, apoptosis, migration and invasion were investigated, and the potential mechanism of lnc-SLC4A1-1 was further explored in BC cells.
Materials and methods
Clinical tissue and blood samples collection
This study obtained ethical approval from the ethics committee of Jiangsu Taizhou People’s Hospital. A total of 66 paired tumor tissues and adjacent normal tissues were collected from BC patients (aged 33–64 years) who underwent surgery in our hospital between October 2015 to October 2018. The specimens were confirmed by HE staining and stored in liquid nitrogen. In addition, 10–15 mL peripheral blood was obtained from BC patients; meanwhile, the same amount of peripheral blood was drawn from 16 paired healthy female subjects (aged 35–62 years). Serum was extracted using density gradient centrifugation and stored at −70 °C. Meanwhile, the clinical information of BC patients, including age, TNM stage, lymph nodes metastasis, estrogen receptor (ER), progesterone receptor (PR), human epidermal growth factor receptor 2 (HER-2), differentiation, tumor size, and survival time, were recorded.
Cell lines
Normal human mammary epithelial cell line MCF10A, as well as human breast cancer cell lines MCF-7 and MDA-MB-231 were obtained from Shanghai Obio Technology Co., Ltd. The cells were maintained in DMEM (Gibco, Carlsbad, CA) containing 10% fetal bovine serum (FBS, Gibco) with standard incubation conditions (5% CO2 and 37 °C).
Cell transfection and treatment
The constructions of lnc-SLC4A1-1 overexpressed vector (pc-lnc-SLC4A1-1) and empty vector (pcDNA-3.1), lnc-SLC4A1-1 silence vector (sh-lnc-SLC4A1-1) and the control vector (sh-NC), as well as CXCL8 overexpressed vector (pEX-CXCL8) and empty vector (pEX-2) were all performed by Biosyntech Co., Ltd. (Suzhou, China). Both MCF-7 and MDA-MB-231 were transfected with pcDNA-3.1, pc-lnc-SLC4A1-1, sh-NC, sh-lnc-SLC4A1-1, combined sh-lnc-SLC4A1-1 + pEX-2, and combined sh-lnc-SLC4A1-1 + pEX-CXCL8, respectively, by Lipofectamine 2000 (Invitrogen, Gaithersburg, MD). To determine the effects of different transfections on the expression of cell migration and invasion related proteins, untreated cells or transfected cells were treated with transforming growth factor (TGF)-β1 (Sigma, St. Louis, MO). In addition, to explore the relationship of lnc-SLC4A1-1 and NF-κB pathway, MCF-7 and MDA-MB-231 were, respectively, treated with NF-κB antagonist (Sigma) and pc-lnc-SLC4A1-1.
Quantitative real-time reverse transcription PCR (qRT-PCR)
Total RNA from tissues and serum was obtained by Trizol (Invitrogen, Gaithersburg, MD). Then cDNA was obtained by reverse transcription of RNA using PrimeScript™ RT reagent Kit (Takara, Dalian, China). The RT-qPCR was performed by a SYBR Premix Ex Taq TM II (Takara) with primers as following: lnc-SLC4A1-1 forward, 3′-CCGTAGACAACTGGACACTCAG-5′ and reverse, 3′-TCATATTCCTCCTGCTCCAGAT-5'; CXCL8 forward, 3′-ACTGAGAGTGATTGAGAGTGGAC-5′ and reverse, 3′-AACCCTCTGCACCCAGTTTTC-5′; Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) forward, 3′-TGTGGGCATCAATGGATTTGG-5′ and reverse 3′-ACACCATGTATTCCGGGTCAAT-5′. GAPDH was served as internal control of measuring lnc-SLC4A1-1 level and data were analyzed with 2−ΔΔCt method.
Chromatin immunoprecipitation (ChIP) assay
Firstly, the mechanism of lnc-SLC4A1-1 was explored by genome bioinformatics analysis (http://genome.ucsc.edu/). Then ChIP assay was performed as previously described [Citation11]. Briefly, the cross-linking of protein–chromatin in cells were performed using 1% formaldehyde, followed by the cross-linking termination using 0.125 M glycine. Subsequently, ultrasonic oscillation of cell lysates was carried out to extract the short fragments of chromatin DNA. After the immunoprecipitation with H3K27ac antibody (Abcam, Cambridge, UK), the chromatin DNA was incubated with protein A/G beads, washed with different buffers: low salt buffers, high salt buffers, LiCl buffers and TE buffer, in turn, and then reverse incubated with proteinase K to induce the decrosslinking. Finally, DNA was purified and used to measure the enrichment of lnc-SLC4A1-1 by qRT-PCR.
MTT assay
The MDA-MB-231 or MCF-7 cells transfected with sh-lnc-SLC4A1-1, pEX-CXCL8 or their corresponding controls were grown in 96-well plates. Then MTT (10 μL, 5 mg/mL) (Sigma) was added into each well every 24 h for 4 h, followed by the addition of 100 μL dimethyl sulfoxide (DMSO, Sigma). The zero hole (medium, MTT, DMSO) and blank hole were set up. The absorbances at 570 nm were read by a microplate reader (Molecular Devices, Sunnyvale, CA).
Flow cytometry analysis
Annexin V-FITC Apoptosis Detection Kit was used to evaluate the cell apoptosis. The MDA-MB-231 or MCF-7 cells with various treatments were digested with Trypsin. After washing with PBS, chondrocytes were resuspended with 1 × Binding Buffer, followed the incubation with PI and FITC-Annexin V for 15 min at 25 °C in the dark, and mixed with 1 × Binding Buffer. Cells were finally detected using flow cytometer (BD, San Jose, CA).
Transwell assay
Transwell inserts (Corning, New York, NY) was used to detect cell migration and invasion abilities. Firstly, the cells with different treatments were plated onto the upper chamber coated with Matrigel containing medium free of serum. Meanwhile, DMEM with 10% FBS was added into the bottom chamber for 48 h. Next, the cells in the bottom chamber insert were treated with 4,6-diamidino-2-phenylindole for 5 min. The number of migrated or invaded cells was calculated by an inverted microscope (Olympus, Tokyo, Japan).
RNA immunoprecipitation (RIP) assay
Cells were treated as ChIP assay, including fixation with formaldehyde and cell lysis. Then cell supernatants were incubated with preprocessed protein-A/G beads by mixing with different antibodies against P50/P105, P65, AP-1 and c-Jun (Cell Signaling Technology, Boston, MA) or IgG control (Millipore, Burlington, MA) at 4 °C overnight. After washing, RNA that co-precipitated was extracted and used for qRT-PCR analysis.
Western blotting
The cells with various treatments were collected, and then lysed by RIPA lysis buffer (Gibco) supplemented with phenylmethanesulfonyl fluoride (PMSF, 1 mM, Sigma). Protein was extracted by centrifugation and detected by the BCA kit (Shanghai Sangon Biotech Co., Ltd, China). Then protein sample was separated on SDS-PAGE gel, and transferred to polyvinylidene fluoride membranes, followed by the blockage with 5% nonfat milk for 1 h. Next, the membrane was incubated with anti-human Bcl-2, Bax, pro-caspase-3, cleaved-caspase-3, pro-caspase-9, cleaved-caspase-9, NF-κB p65, CXCL8, epithelium-cadherin (E-cad), neural-cadherin (N-cad), vimentin (Vim), Snail or β-actin antibody (1:500, Santa Cruz, CA) overnight at 4 °C, and then washed with PBS, followed by the incubation with second antibody (1:5000, Jackson, MI) for 2 h at room temperature, respectively. Lastly, enhanced chemiluminescence (ECL, Millipore) was used to detect the protein levels.
Statistical analysis
GraphPad Prism 6 software was used for data statistical analysis. Enumeration data were expressed as number and analyzed by chi-square test. Measurement data were expressed as the mean ± SD and analyzed by one-way ANOVA followed by multiple comparisons with Tukey test. A value of p < .05 was considered significant and p < .01 was considered highly significant.
Results
Lnc-SLC4A1-1 expression is upregulated in BC tissue, blood samples and cell lines
Compared with adjacent normal tissues, lnc-SLC4A1-1 expression was significantly increased in BC tumor tissues (p < .01, . Similarly, lnc-SLC4A1-1 expression was obviously higher in serum of BC patients than that of health subjects (p < .01, . Then the relationship of lnc-SLC4A1-1 expression and clinicopathological characteristics were investigated. Among the 66 BC patients, 30 were classified as high-expression and the remaining 36 were low-expression. The results showed that BC patients with high-expressed lnc-SLC4A1-1 had a poorer prognosis than those with low-expressed lnc-SLC4A1-1 (p = .021, . In addition, lnc-SLC4A1-1 overexpression was closely associated with higher TNM stage, lymph nodes metastasis, negative PR expression, and higher differentiation (p < .05), while no significant differences were found in age, ER, HER-2, and tumor size between high-expression and low-expression groups (). Consistent with in vivo experiments, in vitro study also found that lnc-SLC4A1-1 expression was conspicuously elevated in MDA-MB-231 (p < .05, ) and MCF-7 cells (p < .01, ) compared with normal MCF 10 A cells.
Figure 1. The expression of lnc-SLC4A1-1 in breast cancer (BC) tissues, blood samples and cell lines. (A) The lnc-SLC4A1-1 level in tumor sample and adjacent normal samples by qRT-PCR. (B) The lnc-SLC4A1-1 level in serum of healthy subjects and BC patients by qRT-PCR. (C) The relationship between lnc-SLC4A1-1 expression and survival times in BC patients. (D) The lnc-SLC4A1-1 level in normal cells (MCF10A) and cancer cells (MDA-MB-231 and MCF-7) by qRT-PCR. *p < .05 and **p < .01 versus Non-tumor, Health, or MCF10A.
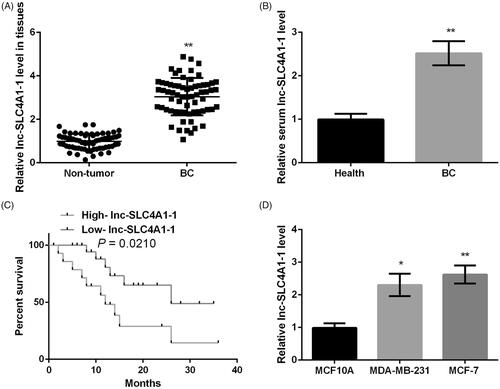
Table 1. Correlation between lnc-SLC4A1-1 expression and clinicopathological characteristics of breast cancer.
Lnc-SLC4A1-1 is transcriptionally activated by H3K27 acetylation in breast cancer
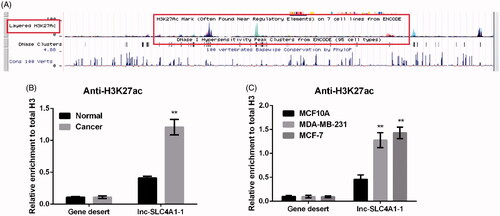
Genome bioinformatics analysis revealed that H3K27ac is highly enriched in the promoter region of lnc-SLC4A1-1 (), indicating that acetylation might be a potential mechanism of lnc-SLC4A1-1. ChIP assay showed that compared with normal tissues and MCF10A cells, H3K27ac enrichment in the promoter region of lnc-SLC4A1-1 was significantly elevated in BC tumor tissues (p < .01, ) as well as MDA-MB-231 and MCF-7 cells (p < .01, ), respectively.
Figure 2. Abnormal expression of lnc-SLC4A1-1 was associated with H3K27 acetylation in breast cancer (BC). (A) Example of promoter region of lnc-SLC4A1-1 in UCSC genome browser. (B) The relationship of lnc-SLC4A1-1 expression and acetylation in BC tissues by chromatin immunoprecipitation (ChIP) assay. (C) The relationship of lnc-SLC4A1-1 expression and acetylation in BC cells by ChIP assay. **p < .01 versus Normal or MCF10A.
Effects of lnc-SLC4A1-1 expression on cell viability, cell apoptosis, migration and invasion
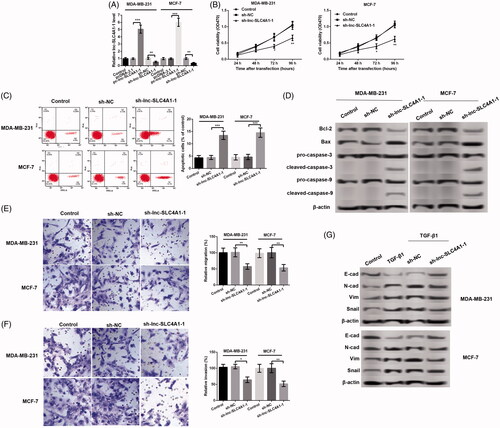
Firstly, both in the MDA-MB-231 and MCF-7 cells, compared with untreated or control cells, lnc-SLC4A1-1 expression was significantly up-regulated in cells with pc-lnc-SLC4A1-1, and remarkably down-regulated after transfection with sh-lnc-SLC4A1-1 (p < .01, . MTT assay showed that lnc-SLC4A1-1 knockdown obviously inhibited cell viability in time-dependent manner both in the MDA-MB-231 and MCF-7 cells (p < .05, . Flow cytometry analysis found that the number of apoptotic cells was prominently increased in cells transfected with sh-lnc-SLC4A1-1 compared with cells with sh-NC in MDA-MB-231 and MCF-7 cells (p < .01, . Consistently, western blotting showed that lnc-SLC4A1-1 knockdown distinctly inhibited Bcl-2 level, as well as, promoted the expression of Bax, cleaved-caspase-3, and cleaved-caspase-9 (. In addition, lnc-SLC4A1-1 knockdown significantly suppressed cell migration (p < .05, ) and invasion (p < .05, ) both in the MDA-MB-231 and MCF-7 cells. Meanwhile, TGF-β1 treatment obviously inhibited E-cad expression and increased the expression of N-cad, Vim, and Snail; while lnc-SLC4A1-1 knockdown counteracted the effects of TGF-β1 on the expression of these migration-related proteins (.
Figure 3. Lnc-SLC4A1-1 knockdown inhibited cell growth in BC cells. (A) lnc-SLC4A1-1 expression of cells transfected with pc-lnc-SLC4A1-1, sh-lnc-SLC4A1-1 or their corresponding control in MDA-MB-231 and MCF-7 cells. (B) Cell viability of untreated cells, cells with sh-NC or sh-lnc-SLC4A1-1 in MDA-MB-231 and MCF-7 cells by MTT assay. (C) The number of apoptotic cells of untreated cells, cells with sh-NC or sh-lnc-SLC4A1-1 in MDA-MB-231 and MCF-7 cells by flow cytometry analysis. (D) The expression of apoptosis-related proteins (Bcl-2, Bax, pro-caspase-3, cleaved-caspase-3, pro-caspase-9, and cleaved-caspase-9) of untreated cells, cells with sh-NC or sh-lnc-SLC4A1-1 in MDA-MB-231 and MCF-7 cells by western blotting. (E) Cell migration of untreated cells, cells with sh-NC or sh-lnc-SLC4A1-1 in MDA-MB-231 and MCF-7 cells by Transwell. (F) Cell invasion of untreated cells, cells with sh-NC or sh-lnc-SLC4A1-1 in MDA-MB-231 and MCF-7 cells by Transwell. (G) The expression of epithelial-mesenchymal transition-related proteins (E-cad, N-cad, Vim, and Snail) of untreated cells, cells with TGF-β1, sh-NC + TGF-β1 or sh-lnc-SLC4A1-1 + TGF-β1 in MDA-MB-231 and MCF-7 cells by western blotting. *p < .05, **p < .01 and ***p < .001.
Lnc-SLC4A1-1 interacts with NF-κB to promote CXCL8 expression
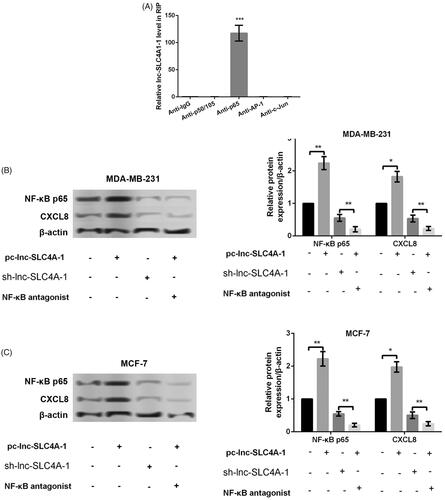
To investigate whether lnc-SLC4A1-1 interacted NF-κB, RIP assay was performed and the results showed that in comparison with p50/105, AP-1 and c-Jun, p65 was significantly enriched (p < .001, ), indicated the specific interaction between lnc-SLC4A1-1 interacted p65. In addition, the further experiments revealed that lnc-SLC4A1-1 overexpression remarkably promoted the expression of NF-κB p65 and CXCL8, while the opposite effects were found after lnc-SLC4A1-1 knockdown (p < .05, . Noteworthily, NF-κB antagonist also obviously inhibited CXCL8 expression in cells with lnc-SLC4A1-1 overexpression (p < .01, . These results suggested that lnc-SLC4A-1 could activate CXCL8 which was dependent on p65.
Figure 4. Lnc-SLC4A1-1 promoted CXCL8 expression. (A) Identification of the transcription factors (P50/P105, P65, AP-1 and c-Jun) related to lnc-SLC4A1-1 by RNA immunoprecipitation assay. (B) The expression of NF-κB p65 and CXCL8 in cells treated with pc-lnc-SLC4A1-1, sh-lnc-SLC4A1-1 or pc-lnc-SLC4A1-1 + NF-κB antagonist in MDA-MB-231 cells by western blotting. (C) The expression of NF-κB p65 and CXCL8 in cells treated with pc-lnc-SLC4A1-1, sh-lnc-SLC4A1-1 or pc-lnc-SLC4A1-1 + NF-κB antagonist in MCF-7 cells by western blotting. *p < .05, **p < .01 and ***p < .001.
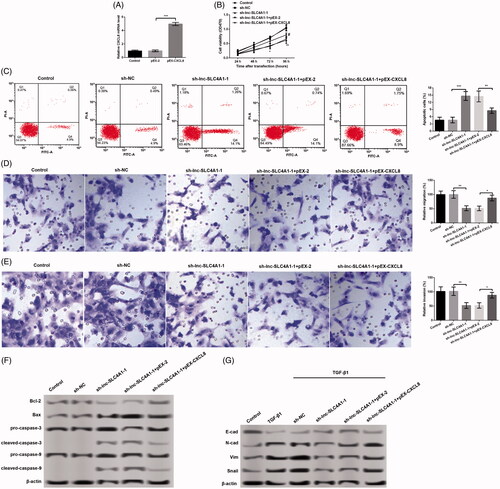
Effects of lnc-SLC4A1-1 on cell viability, cell apoptosis, migration and invasion are associated with CXCL8
To further confirm whether the effects of lnc-SLC4A1-1 on cell viability, apoptosis, migration and invasion were achieved by regulation CXCL8, sh-lnc-SLC4A1-1 and pEX-CXCL8 were co-transfected into MCF-7 cells. The results revealed that the mRNA level of CXCL8 was significantly increased in cells with pEX-CXCL8 compared with cells with pEX-2 (p < .01, . Compared with cells with sh-lnc-SLC4A1-1, the number of apoptotic cells was obviously inhibited (p < .01, ), meanwhile, cell viability (p < .01, ), cell migration (p < .01, ), and cell invasion (p < .01, ) were all conspicuously increased in cells with sh-lnc-SLC4A1-1 + pEX-CXCL8. In addition, the expression of apoptosis-related proteins (Bcl-2, Bax, cleaved-caspase-3, and cleaved-caspase-9) () and epithelial-mesenchymal transition (EMT)-related proteins (E-cad, N-cad, Vim, and Snail) () was consistent with cell apoptosis, migration and invasion after different treatments. These results suggested that CXCL8 overexpression partly reversed the effects of lnc-SLC4A1-1 knockdown on cell viability, apoptosis, migration and invasion.
Figure 5. Relationship of CXCL8 and the effects of lnc-SLC4A1-1 in breast cancer. (A) CXCL8 expression in MCF-7 cells transfected with pEX-2, or pEX-CXCL8. (B) Cell viability of untreated cells, cells with sh-NC, sh-lnc-SLC4A1-1, sh-lnc-SLC4A1-1 + pEX-2, or sh-lnc-SLC4A1-1 + pEX-CXCL8 in MCF-7 cells by MTT assay. (C) The number of apoptotic cells of untreated cells, cells with sh-NC, sh-lnc-SLC4A1-1, sh-lnc-SLC4A1-1 + pEX-2, or sh-lnc-SLC4A1-1 + pEX-CXCL8 in MCF-7 cells by flow cytometry analysis. (D) Cell migration of untreated cells, cells with sh-NC, sh-lnc-SLC4A1-1, sh-lnc-SLC4A1-1 + pEX-2, or sh-lnc-SLC4A1-1 + pEX-CXCL8 in MCF-7 cells by Transwell. (E) Cell invasion of untreated cells, cells with sh-NC, sh-lnc-SLC4A1-1, sh-lnc-SLC4A1-1 + pEX-2, or sh-lnc-SLC4A1-1 + pEX-CXCL8 in MCF-7 cells by Transwell. (F) The expression of apoptosis-related proteins (Bcl-2, Bax, pro-caspase-3, cleaved-caspase-3, pro-caspase-9, and cleaved-caspase-9) of untreated cells, cells with sh-NC, sh-lnc-SLC4A1-1, sh-lnc-SLC4A1-1 + pEX-2, or sh-lnc-SLC4A1-1 + pEX-CXCL8 in MCF-7 cells by western blotting. (G) The expression of epithelial-mesenchymal transition-related proteins (E-cad, N-cad, Vim and Snail) of untreated cells, cells with sh-NC, sh-lnc-SLC4A1-1, sh-lnc-SLC4A1-1 + pEX-2, or sh-lnc-SLC4A1-1 + pEX-CXCL8 in MCF-7 cells by western blotting. *p < .05, **p < .01 and ***p < .001.
Discussion
In the current study, lnc-SLC4A1-1 was identified as significantly up-regulated in BC tissue, serum samples and cell lines. In BC cells, lnc-SLC4A1-1 knockdown significantly promoted cell apoptosis and suppressed cell proliferation, migration and invasion. Furthermore, the results showed that elevated H3K27 acetylation in the promoter region of lnc-SLC4A1-1 activated transcription of lnc-SLC4A1-1, and lnc-SLC4A1-1 could interact with transcription factor NF-κB p65, thereby regulating CXCL8 expression. Meanwhile, this study found that the effects of lnc-SLC4A1-1 knockdown on BC cells could be partly reversed by CXCL8 overexpression.
Currently, numerous studies have suggested that lncRNAs play important roles in cancer development and can be considered as biomarkers responsible for cancer diagnosis and treatment. This study preliminarily focused on the effects of lnc-SLC4A1-1 that newly discovered and identified as overexpression in unexplained recurrent pregnancy loss [Citation10] on BC. The results found that lnc-SLC4A1-1 was overexpressed in BC, and lnc-SLC4A1-1 knockdown inhibited cell viability and promoted cell apoptosis in both MDA-MB-231 and MCF-7 cells. Meanwhile, apoptosis-related proteins were detected, and lnc-SLC4A1-1 knockdown distinctly inhibited Bcl-2 level as well as promoted the expression of Bax, cleaved-caspase-3, and cleaved-caspase-9. It is well known that the mitochondria-mediated apoptosis pathway is a classical pathway to contribute to cell apoptosis in cancers [Citation12,Citation13]. Caspase-3 as a downstream molecule can induce cell apoptosis by regulating the ratio of Bcl-2/Bax [Citation13,Citation14]. In addition, the recruitment and activation of caspase-9 also can subsequently activate caspase 3 [Citation12]. These results suggested that the effect of lnc-SLC4A1-1 on cell apoptosis was dependent on classical mitochondria-mediated apoptosis pathway. Moreover, this study revealed decreased cell migration and invasion induced by lnc-SLC4A1-1 knockdown in MDA-MB-231 and MCF-7 cells. EMT has been evidenced to contribute to BC metastasis by activating TGF-β1 signaling pathway [Citation15]. During EMT, E-cad expression is significantly inhibited, while the expression of N-cad, Vim, and Snail is obviously elevated [Citation16]. Consistently, this study showed the opposite expression trend of E-cad, N-cad, Vim, and Snail induced by lnc-SLC4A1-1 knockdown, indicating that the effects of lnc-SLC4A1-1 on BC cell migration and invasion was mediated by EMT. Collectively, this study prompted that lnc-SLC4A1-1 was a potential tumor promoter in BC.
Furthermore, the mechanism of lnc-SLC4A1-1 in BC was investigated. Recent studies have demonstrated that several lncRNAs which originated from the enhancer regions can regulate gene expression by interacting with transcription factors [Citation17–19]. This study found high H3K27ac enrichment in the promoter region of lnc-SLC4A1-1 in BC tissues and cells, and it is reported that high H3K27ac enrichment usually appears in enhancer regions [Citation20]. Thus, we speculated that lnc-SLC4A1-1 might be transcribed from enhancer regions with high H3K27ac enrichment, which also has been confirmed by a previous study [Citation10]. This study further revealed that lnc-SLC4A1-1 could interact with NF-κB p65. NF-κB signaling pathway has been reported to be related to participate in the growth and metastasis of tumor cells [Citation21]. The previous study has shown that NF-κB activation can regulate the expression of Bcl-2 family to induce cell apoptosis [Citation22]. Notably, NF-κB activation is reported to play key role in cell proliferation and apoptosis in human BC [Citation23]. In addition, the promoting effect of NF-κB activation on cell migration and invasion is also evidenced to be related to EMF in cancers [Citation24–26]. This study showed that lnc-SLC4A1-1 overexpression remarkably promoted NF-κB p65 expression, which suggested that the tumor promoting effect of lnc-SLC4A1-1 on BC cells might be associated with NF-κB activation.
This study also found that up-regulated lnc-SLC4A1-1 induced CXCL8 overexpression, while NF-κB antagonist inhibited CXCL8 expression, suggested lnc-SLC4A-1 could activate CXCL8 which was dependent on NF-κB. Consistent with our study, Collins et al. have also revealed that IL-8 is transcriptional regulated by NF-κB [Citation27]. CXCL8 (also known as interleukin 8) is involved in inflammatory diseases [Citation28] and cancers [Citation29]. It has been reported that CXCL8 is overexpressed in BC tissues, and can be used as an indicator to distinguish between benign and malignant breast tumors [Citation30]. Several studies have shown that the metastatic activity of malignancy can be induced by CXCL8 by increasing the expression of MMP-2 and MMP-9 [Citation29,Citation31,Citation32]. Another study also has found that CXCL8 is related to the growth of triple-negative BC cells [Citation33]. The present study found that CXCL8 overexpression partly reversed the tumor-inhibiting effect of lnc-SLC4A1-1 knockdown on BC cells, which indicated that the tumor-promoting effect of lnc-SLC4A1-1 might be associated with NF-κB/CXCL8 in BC cells.
In conclusion, findings from our work suggested that lnc-SLC4A1-1 was activated by H3K27ac acetylation, which in turn promotes NF-κB/CXCL8, indicating that lnc-SLC4A1-1 could promote the development of BC by regulating NF-κB/CXCL8.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Senkus E, Kyriakides S, Ohno S, et al. Primary breast cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2015;26(5):v8–v30.
- Sharma R. Breast cancer incidence, mortality and mortality-to-incidence ratio (MIR) are associated with human development, 1990–2016: evidence from Global Burden of Disease Study 2016. Breast Cancer. 2019;26(4):1–18.
- Gradishar WJ, Anderson BO, Balassanian R, et al. NCCN guidelines insights: breast cancer, version 1.2017. J Natl Compr Canc Netw. 2017;15(4):433–451.
- Gradishar WJ, Anderson BO, Balassanian R, et al. Invasive breast cancer version 1.2016, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw. 2016;14(3):324–354.
- Anastasiadou E, Jacob LS, Slack FJ. Non-coding RNA networks in cancer. Nat Rev Cancer. 2018;18(1):5.
- Iyer MK, Niknafs YS, Malik R, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47(3):199.
- Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: a new paradigm. Cancer Res. 2017;77(15):3965–3981.
- Banerjee S, Chakravarti D, Ganguli S, et al. Cancer and noncoding RNAs. Beijing, China: Elsevier; 2018. Chapter 25, Cancer noncoding RNA discovery through high-throughput sequencing; p. 463–477.
- Jiang Y-Z, Liu Y-R, Xu X-E, et al. Transcriptome analysis of triple-negative breast cancer reveals an integrated mRNA-lncRNA signature with predictive and prognostic value. Cancer Res. 2016;76(8):2105–2114.
- Huang Z, Du G, Huang X, et al. The enhancer RNA lnc-SLC4A1-1 epigenetically regulates unexplained recurrent pregnancy loss (URPL) by activating CXCL8 and NF-kB pathway. EBioMedicine. 2018;38:162–170.
- Qin Y, Roberts JD, Grimm SA, et al. An obesity-associated gut microbiome reprograms the intestinal epigenome and leads to altered colonic gene expression. Genome Biol. 2018;19(1):7.
- Lopez J, Tait S. Mitochondrial apoptosis: killing cancer using the enemy within. Br J Cancer. 2015;112(6):957.
- Ichim G, Tait SW. A fate worse than death: apoptosis as an oncogenic process. Nat Rev Cancer. 2016;16(8):539.
- Hatok J, Racay P. Bcl-2 family proteins: master regulators of cell survival. Biomol Concepts. 2016;7(4):259–270.
- Ye X, Brabletz T, Kang Y, et al. Upholding a role for EMT in breast cancer metastasis. Nature. 2017;547(7661):E1.
- Brabletz T, Kalluri R, Nieto MA, et al. EMT in cancer. Nat Rev Cancer. 2018;18(2):128.
- Yang Y, Su Z, Song X, et al. Enhancer RNA-driven looping enhances the transcription of the long noncoding RNA DHRS4-AS1, a controller of the DHRS4 gene cluster. Sci Rep. 2016;6(1):20961.
- Tsai P-F, Dell’Orso S, Rodriguez J, et al. A muscle-specific enhancer RNA mediates cohesin recruitment and regulates transcription in trans. Molecular cell. 2018;71(1):129.e8–141.e8.
- Léveillé N, Melo CA, Rooijers K, et al. Genome-wide profiling of p53-regulated enhancer RNAs uncovers a subset of enhancers controlled by a lncRNA. Nat Commun. 2015;6(1):6520.
- Creyghton MP, Cheng AW, Welstead GG, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci. 2010;107(50):21931–21936.
- Taniguchi K, Karin M. NF-κB, inflammation, immunity and cancer: coming of age. Nat Rev Immunol. 2018;18(5):309.
- Karin M, Lin A. NF-kappaB at the crossroads of life and death. Nat Immunol. 2002;3(3):221.
- Biswas DK, Shi Q, Baily S, et al. NF-κB activation in human breast cancer specimens and its role in cell proliferation and apoptosis. Proc Natl Acad Sci. 2004;101(27):10137–10142.
- Lin Z, Li W, Zhang H, et al. CCL18/PITPNM3 enhances migration, invasion, and EMT through the NF-κB signaling pathway in hepatocellular carcinoma. Tumor Biol. 2016;37(3):3461–3468.
- Gu K, Li M-M, Shen J, et al. Interleukin-17-induced EMT promotes lung cancer cell migration and invasion via NF-κB/ZEB1 signal pathway. Am J Cancer Res. 2015;5(3):1169.
- Wu Y-D, Zhou B. TNF-alpha/NF-kappaB/snail pathway in cancer cell migration and invasion. Br J Cancer. 2010;102(4):639.
- Collins TS, Lee L-F, Ting J-Y. Paclitaxel up-regulates interleukin-8 synthesis in human lung carcinoma through an NF-κB-and AP-1-dependent mechanism. Cancer Immunol, Immunother. 2000;49(2):78–84.
- Russo RC, Garcia CC, Teixeira MM, et al. The CXCL8/IL-8 chemokine family and its receptors in inflammatory diseases. Exp Rev Clin Immunol. 2014;10(5):593–619.
- Liu Q, Li A, Tian Y, et al. The CXCL8-CXCR1/2 pathways in cancer. Cytokine Growth Factor Rev. 2016;31:61–71.
- Todorović-Raković N, Milovanović J. Interleukin-8 in breast cancer progression. J Interferon Cytokine Res. 2013;33(10):563–570.
- Ma Y, Ren Y, Dai Z-J, et al. IL-6, IL-8 and TNF-α levels correlate with disease stage in breast cancer patients. Adv Clin Exp Med. 2017;26(3):421–426.
- Chen C, Duckworth C, Fu B, et al. Circulating galectins -2, -4 and -8 in cancer patients make important contributions to the increased circulation of several cytokines and chemokines that promote angiogenesis and metastasis. Br J Cancer. 2014;110(3):741.
- Hartman ZC, Poage GM, Den Hollander P, et al. Growth of triple-negative breast cancer cells relies upon coordinate autocrine expression of the proinflammatory cytokines IL-6 and IL-8. Cancer Res. 2013;73(11):3470–3480.
