Abstract
Aims
In previous studies, numerous differential lncRNAs in cerebral ischemic reperfusion injury were identified using RNA-Seq analysis. However, little is known about whether and how lncRNAs involved in cerebral I/R injury. In this study, we investigated the function and explored the possible mechanism of lncRNA Gm11974 in cerebral I/R injury.
Methods
Oxygen glucose deprivation model in N2a cells were utilized to mimic the cerebral I/R injury in vitro. Trypan blue staining, Tunel, JC-1 and cell viability were measured to evaluate the function of lncRNA Gm11974. Dual-luciferase reporter assay was used to explore the potential mechanism of lncRNA Gm11974.
Results
Gm11974 was mainly located in cytoplasm. Knockdown of lncRNA Gm11974 alleviated the apoptosis induced by OGD and cell death rates were significantly reduced. We further provided the possible mechanism that Gm11974/miR-766-3p/NR3C2 axis plays important role in cerebral I/R injury.
Conclusions
We evaluated the function and mechanism of lncRNA Gm11974 in ischemic brain injury. LncRNA Gm11974 may serve as a potential target for new therapeutic intervention.
Introduction
China bears the biggest stroke burden in the world. And the prevalence of stroke is keeping increasing over past 30 years, especially in rural areas [Citation1]. It is reported that the age-standardized prevalence, incidence, and mortality rates were 1114.8/100000 people, 246.8 and 114.8/100000 person-years [Citation2,Citation3]. Stroke is defined as a medical condition where poor blood flow to the brain results in cell death and symptoms like lose ability to move or feel one side of the body, to speak or to understand or loss the vision of one side [Citation2]. The pathological changes of stroke are usually characterized by cerebral ischemia-reperfusion injury followed by cells swelling and apoptosis [Citation4,Citation5]. A great number of researches have been carried out to reveal the inner internal mechanism of cerebral ischemia-reperfusion injury [Citation6]. However, because of the heterogeneity, it is hard to draw any conclusions and the mechanism of cerebral ischemia-reperfusion injury remains unclear.
Non-coding RNAs (ncRNAs) are the transcripts that cannot be translated in proteins [Citation7]. Non-coding RNAs include transfer RNAs (tRNAs), ribosomal RNAs (rRNAs), microRNAs, siRNAs, long non-coding RNAs (lncRNA) and so on [Citation8–10]. Among these ncRNAs, microRNA and lncRNA have been found playing a significant role in cerebral ischemia-reperfusion injury [Citation11]. For example, Gai HY and his colleagues demonstrated that long non-coding RNA (lncRNA) cardiac hypertrophy-related factor (CHRF) could function as a competing endogenous RNA (ceRNA) and compete with Sex-determining region Y box 6 (SOX6) to direct binding with miR-126 and further regulate the progress of cerebral ischemia-reperfusion injury [Citation12]. Wei R et al. found long non-coding RNA AK038897 could aggravate cerebral ischemia/reperfusion injury via acting as a ceRNA for miR-26a-5p to target DAPK1 [Citation13]. Zhong Y et al. found lncRNA SNHG14 promotes inflammatory response induced by cerebral ischemia/reperfusion injury through regulating miR-136-5p/ROCK1 [Citation14]. In current research, we demonstrated the up-regulation of lncRNA Gm11974 in oxygen-glucose deprivation (OGD) cell line and found the expressions of lncRNA Gm11974 is correlated with duration of OGD which indicated that lncRNA Gm11974 may play a role the progress of cerebral ischemia/reperfusion injury.
We investigated the function and explored the possible mechanism of lncRNA Gm11974 in cerebral I/R injury. Oxygen glucose deprivation model in N2a cells were utilized to mimic the cerebral I/R injury in vitro. Trypan blue staining, Tunel, JC-1 and cell viability were measured to evaluate the function of lncRNA Gm11974. Our results showed that lncRNA Gm11974 was mainly located in cytoplasm. Knockdown of lncRNA Gm11974 alleviated the apoptosis induced by OGD and cell death rates were significantly reduced. We further provided the possible mechanism that Gm11974/miR-766-3p/NR3C2 axis plays important role in cerebral I/R injury. We evaluated the function and mechanism of lncRNA Gm11974 in ischemic brain injury. LncRNA Gm11974 may serve as a potential target for new therapeutic intervention.
Materials and methods
Cell culture
HEK293T and N2a cells were purchased from Shanghai Institute of Academy. HEK293T and N2a cells were cultured in Dulbecco's Modification of Eagle's medium (DMEM; Gibco) containing 10% fetal bovine serum (FBS; Gibco) in 5% CO2 at 37 °C.
Plasmid transfection
The knockdown siRNA lncRNA Gm11974 were constructed by Gene Pharma (Shanghai, China). Overexpression and knockdown vector miR-766-3p were purchased from Gene Pharma (Shanghai, China). The 3′-untranslated region (UTR) of NR3C2, with wild-type (WT) or mutant (mut) binding sites for miR-766-3p, was amplified and cloned into the pGL3 vector (Promega, Madison, WI) for dual-luciferase reporter assay. For transfection, HEK293T and N2a cells were transfected with different vectors as indicated in figures by lipo2000 (Invitrogen) following the protocols when cells confluence reached 60%. After 36 h transfection, the cells were harvested for further experiment.
OGD exposure
Oxygen glucose deprivation model was performed as described previously [Citation15]. Briefly, N2a cells were cultured in serum-free and glucose-free medium under 95% N2 and 5% CO2 at 37 °C for 3 h. After 3 h culture, the medium was replaced with normal medium for different time to mimic the reperfusion injury.
Cell viability
N2a cells were seed into 96-well plates with 2000 cells each well. Each well was added 10 μL 3‐(4,5‐dimethylthiazol‐2‐yl)‐2,5‐diphenyltetrazolium bromide (MTT) solution (5 mg/mL, Beyotime, Shanghai, China) and incubated for 4 h at 37 °C. Cell viability was measured by a microplate reader at 570 nm (Bio‐Tek, Winooski, VT).
Death rates
After 2 min digestion, the cells were pipette thoroughly. Trypan blue staining was used to evaluate the cell death rates.
Tunel assay
Terminal deoxynucleotidy1 transferase-mediated dUTP nick end-labeling (TUNEL) assay were performed to evaluate the apoptosis effect of lncRNA Gm11974. After OGD exposure, N2a cells were incubated with TdT and fluorescein-labelled dUTP for 45 min at 37 °C. Then FACSCalibur flow cytometer was used to measure the percentage of apoptotic of cells.
JC-1 fluorescence
After OGD exposure, JC-1 at a concentration of 10 μg/ml (Beyotime, C2006, Shanghai, China) was added to the medium for 10 min. N2a cells were then washed twice in PBS. Fluorescence emission was recorded at 485 and 580 nm.
Real-time PCR
Total RNA was isolated from cultured N2a cell and HEK293T cells by Trizol Reagent (Thermo Fisher Scientific, MA, USA), respectively. Quantitative analysis of qRT-PCR was carried out using SYBR Premix Ex TaqTM (TaKaRa, Dalian, China). Relative expression levels of lncRNA Gm11974, miR-766-3p, NR3C3 and GAPDH mRNA were calculated by using the 2 − ΔΔCT method.
Western blot
HEK293T and N2a cells were lysed by RIPA lysis containing proteinase inhibitor (Roche, USA). BCA protein assay kit (Pierce, Rockford, IL, USA) was used to calculate the total protein concentrations. Protein samples (40 μg) were resolved by 10% SDS-PAGE gel and transferred to polyvinylidene difluoride membrane (Roche, USA). After 2 h blocking, membranes were incubated against primary antibodies Bax (1:1000, CST), Bcl-2 (1:1000, CST), NR3C2(1:1000, CST) and GAPDH (1:5000, CST) at 4 °C overnight, followed by incubation with a peroxidase-conjugated antibody. Immunopositively bands were analyzed using a FluorChem M system (ProteinSimple, San Jose, CA, USA).
Dual-luciferase reporter assay
The 3′-UTR of NR3C2, with wild-type or mutant (mut) binding sites for miR-766-3p, was amplified and cloned into the pGL3 vector (Promega, Madison, WI, USA) to generate the plasmid pGL3-wt-NR3C2-3′-UTR or pGL3-mut-NR3C2-3′-UTR. HEK 293 T cells were used to perform the luciferase reported assay, and the miR-766-3p mimics or inhibitor and the NR3C2 vector were transfected using Lipofectamine 2000 reagent. Luciferase activity was analyzed using Dual-Luciferase system following the manufacture’s protocol.
Statistical analysis
SPSS 23.0 was used to calculate the values (means ± standard error of the mean). Statistical analyses were analyzed using two-sided Student’s t-test or one-way ANOVA. The statistical significance was p < .05.
Results
OGD exposure and preliminary analysis of lncRNA Gm11974
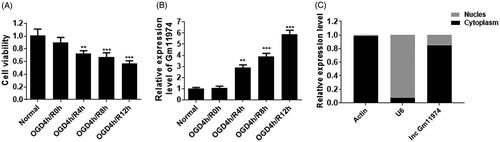
To establish the OGD model, firstly, we used serum-free and glucose-free medium to mimic the cerebral ischemic reperfusion injury. After 4 h OGD exposure, we treated cells with different time course of reperfusion. The cell viability was significantly decreased with the time development (. At the same time, we detected the expression of lncRNA Gm11974 under the OGD exposure. The expression of lncRNA Gm11974 was significantly increased with the time development and reached the highest changes in the 12 h reperfusion (. In addition, we also detected the location of lncRNA Gm11974. As shown in , lncRNA Gm11974 was mainly located in cytoplasm.
Function of lncRNA Gm11974 in cerebral ischemic reperfusion injury
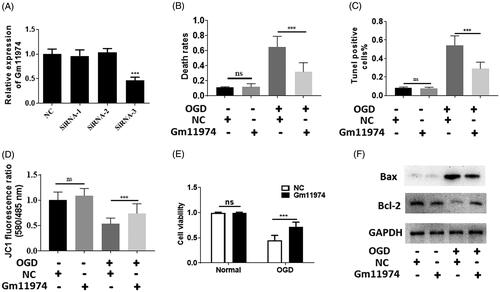
To investigate the function of lncRNA Gm11974in cerebral ischemic reperfusion injury, firstly, we used siRNAs to knockdown the lncRNA Gm11974. As shown in , the expression of lncRNA Gm11974 was downregulated using siRNA-3. Next, we performed comprehensive analysis to assess the function of lncRNA Gm11974. Trypan blue staining results showed that knockdown of lncRNA Gm11974 significantly decreased the cell death rates (. We also performed the tunel assay to detect the cell apoptosis effect. Knockdown of lncRNA Gm11974 significantly reduced the cell apoptosis (). Mitochondrial membrane potential was measured to detect the effect of lncRNA Gm11974. And knockdown of lncRNA Gm11974 protect the mitochondrial membrane potential under OGD exposure (). In addition, MTT results showed that knockdown of lncRNA Gm11974 increased the cell viability (). The protein level of Bax was decreased after knockdown of lncRNA Gm11974 and Bcl-2 was increased, whereas no significant difference was detected under normal conditions (). Thus, our results showed that knockdown of lncRNA Gm11974 can protect N2a cells from cerebral ischemic reperfusion injury and lncRNA Gm11974 possessed a pro-apoptosis effect.
Figure 2. Function of lncRNA Gm11974 in cerebral ischemic reperfusion injury A. Knockdown of lncRNA Gm11974 was verified via real-time PCR. B. Knockdown of lncRNA Gm11974 significantly reduced the cell death rates. C. Tunel results suggested that knockdown of lncRNA Gm11974 significantly reduced the apoptosis cells. D. JC-1 results suggested that knockdown of lncRNA Gm11974 protect mitochondrial membrane potential. E. Cell viability revealed that knockdown of lncRNA Gm11974 significantly increased the cell viability. F Western blotting results revealed that Bax was decreased in the group of knockdowns of lncRNA Gm11974 and Bcl-2 was increased compared with NC group.
LncRNA Gm11974 negatively regulate miR-766-3p
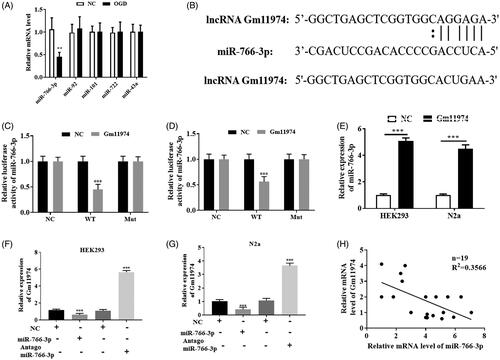
To investigate the mechanism of lncRNA Gm11974 involved in the cerebral ischemic reperfusion injury, we used bio-informatics software to predict whether miRNAs are involved in the regulation. Bio-informatics results predicted that miR-766-3p, miR-92, miR-101, miR-722 and miR-43a may be the potential binding targets (). We then treated N2a cells with OGD and then detect the expression of those miRNAs. Only miR-766-3p was significantly decreased in the OGD exposure. As shown in , the potential binding sequence between miR-766-3p and lncRNA Gm11974 was showed. Next, we used dual-luciferase reporter assay to detect whether lncRNA Gm11974 can regulate miR-766-3p. Our results showed that the relative activity of miR-766-3p was decreased in wt lncRNA Gm11974 group in HEK293T and N2a cells, and there is no significant difference in mut lncRNA Gm11974 group (. Knockdown of lncRNA Gm11974 significantly increased the expression of miR-766-3p in HEK293T and N2a cells (). Overexpression of miR-766-3p manifestly reduced the expression of lncRNA Gm11974 and down-regulated of miR-766-3p distinctly increased the expression of lncRNA Gm11974 (. Our results demonstrated that lncRNA Gm11974 could negatively regulate the expression of miR-766-3p ().
Figure 3. LncRNA Gm11974 negatively regulate miR-766-3p A. Bio-informatics analysis results showed that the potential binding miRNAs. B. The potential binding sequence between lncRNA Gm11974 and miR-766-3p. C. Dual-luciferase reporter assay revealed that lncRNA Gm11974 negatively regulate miR-766-3p in HEK293T cells. D. Dual-luciferase reporter assay revealed that lncRNA Gm11974 negatively regulate miR-766-3p in N2a cells. E. The expression of miR-766-3p was increased after knockdown of lncRNA Gm11974. F. Overexpression of miR-766-3p significantly decreased the expression of lncRNA Gm11974 in HEK293T cells and vice versa. G. Overexpression of miR-766-3p significantly decreased the expression of lncRNA Gm11974 in N2a cells and vice versa. H. LncRNA Gm11974 negatively regulate miR-766-3p.
MiRNA-766-3p negatively regulate NR3C2
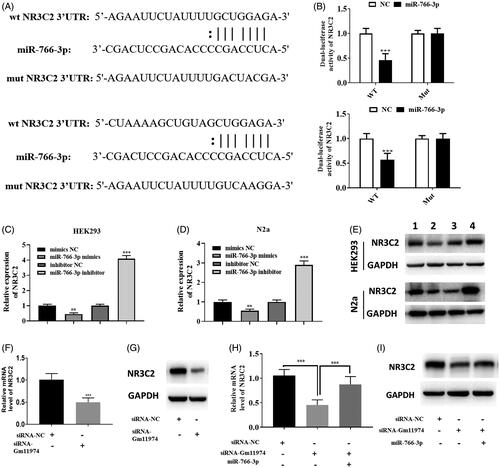
In previous studies, miR-766-3p was reported to regulate NR3C2. In our study, we used bio-informatics analysis to assess whether miR-766-3p could regulate NR3C2. As shown in , the potential binding sequence between miR-766-3p and NR3C2 was showed. Next, dual-luciferase reporter assay showed that miR-766-3p can reduced the activity of NR3C2 in HEK293T and N2a cells, and there are no significant changes in the mut NR3C2 group (). Thus, our results demonstrate that miR-766-3p could regulate NR3C3 in cerebral cells. We performed real-time PCR and western blot to assess the relationship between miR-766-3p and NR3C2. Overexpression of miR-766-3p could significantly reduce the expression of NR3C2 and down-regulated of miR-766-3p could increase the expression of NR3C2 in HEK293T and N2a cells (, ). Similar results could be achieved via western blot, suggesting miR-766-3p could regulate the protein level of NR3C2 (). Thus, we wondered the relationship between lncRNA Gm11974 and NR3C2. Knockdown of lncRNA Gm11974 significantly reduced the expression of NR3C2 via real-time PCR (). Similar results could be achieved via western blot (). And overexpression of miR-766-3p can increased the expression of NR3C2, suggesting that overexpression of miR-766-3p can block the effect of lncRNA Gm11974 (. Thus, our results suggested that miR-766-3p negatively regulate NR3C2.
Figure 4. MiRNA-766-3p negatively regulate NR3C2 A. The potential binding sequence between miR-766-3p and NR3C3. B. Dual-luciferase reporter assay suggested that miR-766-3p significantly reduced the activity of WT NR3C2 in HEK293T cells and N2a cells and had no changes in mut group. C. Overexpression of miR-766-3p significantly reduced the expression of NR3C2 and vice versa in HEK293T cells. D. Overexpression of miR-766-3p significantly reduced the expression of NR3C2 and vice versa in N2a cells. E. Overexpression of miR-766-3p significantly reduced the protein level of NR3C2 and vice versa in HEK293T and N2a cells via western blot. F. The expression of NR3C2 was decreased in the group of knockdowns of lncRNA Gm11974. G. The protein level of NR3C2 was decreased in the group of knockdowns of lncRNA Gm11974. H. Overexpression of miR-766-3p could rescued the effect of lncRNA Gm11974. I. Overexpression of miR-766-3p could rescued the effect of lncRNA Gm11974 via western blot.
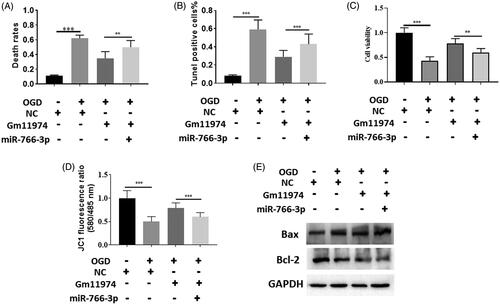
LncRNA Gm11974/miR-766-3p/NR3C2 axis in cerebral ischemic reperfusion injury
To further prove the relationship between lncRNA Gm11974, miR-766-3p and NR3C2, we performed rescue experiment to identify the relationship. Overexpression of miR-766-3p could increase the cell death rates compared with lncRNA Gm11974 group (). Tunel assay showed that co-transfection of lncRNA Gm11974 and miR-766-3p could restored the effect of lncRNA Gm11974 (). Cell viability showed that co-transfection of lncRNA Gm11974 and miR-766-3p decreased the cell viability rate (). In addition, overexpression of miR-766-3p could rescued the effect of lncRNA Gm11974 (). Apoptosis markers, Bax and Bcl-2, were detected via western blot. Co-transfection of lncRNA Gm11974 and miR-766-3p could increase the Bax protein level and decrease the Bcl-2 level (. To date, our results provide the evidence of lncRNA Gm11974/miR-766-3p/NR3C2 axis play an important role in the cerebral ischemic reperfusion injury.
Figure 5. LncRNA Gm11974/miR-766-3p/NR3C2 axis in cerebral ischemic reperfusion injury. A. Overexpression of miR-766-3p could increase the cell death rates compared with lncRNA Gm11974 group. B. Tunel assay showed that co-transfection of lncRNA Gm11974 and miR-766-3p could restored the effect of lncRNA Gm11974. C. Cell viability showed that co-transfection of lncRNA Gm11974 and miR-766-3p decreased the cell viability rate. D. Overexpression of miR-766-3p could rescued the effect of lncRNA Gm11974. E. Bax and Bcl-2 were detected via western blot.
Discussion
In previous studies, numerous differential lncRNAs in cerebral ischemic reperfusion injury were identified using RNA-Seq analysis [Citation16]. In this study, we investigated the function and explored the possible mechanism of lncRNA Gm11974 in cerebral I/R injury. Oxygen glucose deprivation model in N2a cells were utilized to mimic the cerebral I/R injury in vitro. Trypan blue staining, Tunel, JC-1 and cell viability were measured to evaluate the function of lncRNA Gm11974. Our results showed that lncRNA Gm11974 was mainly located in cytoplasm. Knockdown of lncRNA Gm11974 alleviated the apoptosis induced by OGD and cell death rates were significantly reduced. We further provided the possible mechanism that Gm11974/miR-766-3p/NR3C2 axis plays important role in cerebral I/R injury. We evaluated the function and mechanism of lncRNA Gm11974 in ischemic brain injury. LncRNA Gm11974 may serve as a potential target for new therapeutic intervention.
Great efforts have been put into the field of long non coding RNA [Citation17,Citation18]. Long non coding RNA has been widely studied in different biological process, including cell cycle [Citation19], apoptosis [Citation20], RNA splicing [Citation21], translation regulation [Citation22] and ubiquitination [Citation23]. For example, Lnc-CCDST promotes DHX9 degradation through the ubiquitin proteasome pathway by enhancing MDM2-DHX9 interaction [Citation24]. LncRNA PVT1 promotes angiogenesis via activating the STAT3/VEGFA axis in gastric cancer [Citation25]. In our study, we proved that lncRNA Gm11974 negatively regulate the expression of miR-766-3p and positively regulate the protein level of NR3C2. Whether lncRNA Gm11974 could regulate the ubiquitination of NR3C2 remains unknown. Although we proved that knockdown of lncRNA Gm11974 can reduce the protein level of NR3C2, the evidence is not direct and further CO-IP assay should be performed to verify the hypothesis.
To further verify the role of lncRNA Gm11974/miR-766-3p/NR3C2 axis in cerebral ischemic reperfusion injury, we performed comprehensive rescue experiment. We found that expression of NR3C2 was decreased in lncRNA Gm11974 knockdown group. We then overexpressed miR-766-3p in the knockdown group of lncRNA Gm11974. Our results showed that overexpression of miR-766-3p could block the effect of lncRNA Gm11974. Tunel assay showed that co-transfection of lncRNA Gm11974 and miR-766-3p could restored the effect of lncRNA Gm11974. Cell viability showed that co-transfection of lncRNA Gm11974 and miR-766-3p decreased the cell viability rate. As a consequence, the results of rescue experiment are in accordance with our hypothesis and it seems quite reliable to believe that knockdown of lncRNA Gm11974 could alleviate the cell apoptosis by sponging miR-766-3p and inhibiting its function of antagonizing NR3C2.
In summary, we verified the role and function of lncRNA Gm11974 in cerebral stroke and proved its mechanism, which regulate the miR-766-3p and NR3C2. We conducted several comprehensive experiments with the hope to explore the inner association between lncRNA Gm11974 and cerebral ischemia/reperfusion injury and to demonstrate that lncRNA Gm11974 could antagonize miR-776-3p by sponging miR-776-3p who can inhibit the transcription of NR3C2 and further promote the progress of cerebral ischemia/reperfusion injury. The current research may explore the new target for inhibiting or reversing cerebral ischemia/reperfusion injury.
Disclosure statement
All authors declare no conflicts of interest.
Additional information
Funding
References
- Prabhakaran S, Ruff I, Bernstein RA. Acute stroke intervention: a systematic review [J]. JAMA. 2015;313(14):1451–1462.
- Campbell BC, Mitchell PJ, Kleinig TJ, et al. Endovascular therapy for ischemic stroke with perfusion-imaging selection [J]. N Engl J Med. 2015;372(11):1009–1018.
- Jovin TG, Chamorro A, Cobo E, et al. Thrombectomy within 8 hours after symptom onset in ischemic stroke [J]. N Engl J Med. 2015;372(24):2296.
- Ritzel RM, Lai YJ, Crapser JD, et al. Aging alters the immunological response to ischemic stroke [J]. Acta Neuropathol. 2018;136(1):89.
- Jiang X, Andjelkovic AV, Zhu L, et al. Blood-brain barrier dysfunction and recovery after ischemic stroke [J]. Prog Neurobiol. 2018, 163–164:144–171.
- Dyson A, Dal-Pizzol F, Sabbatini G, et al. Ammonium tetrathiomolybdate following ischemia/reperfusion injury: chemistry, pharmacology, and impact of a new class of sulfide donor in preclinical injury models [J]. PLoS Med. 2017;14(7):e1002310.
- Fang W, Shu S, Yongmei L, et al. miR-224-3p inhibits autophagy in cervical cancer cells by targeting FIP200 [J]. Sci Rep. 2016;6:33229.
- Xu R, Feng F, Yu X, et al. LncRNA SNHG4 promotes tumour growth by sponging miR-224-3p and predicts poor survival and recurrence in human osteosarcoma [J]. Cell Prolif. 2018;51(6):e12515.
- Bhalala OG, Srikanth M, Kessler JA. The emerging roles of microRNAs in CNS injuries [J]. Nat Rev Neurol. 2013;9(6):328–339.
- Yang P, Chen T, Xu Z, et al. Long noncoding RNA GAPLINC promotes invasion in colorectal cancer by targeting SNAI2 through binding with PSF and NONO [J]. Oncotarget. 2016;7(27):42183–42194.
- Li EY, Zhao PJ, Jian J, et al. LncRNA MIAT overexpression reduced neuron apoptosis in a neonatal rat model of hypoxic-ischemic injury through miR-211/GDNF [J]. Cell Cycle. 2019 Jan;18(2):156–166.
- Gai HY, Wu C, Zhang Y, et al. Long non-coding RNA CHRF modulates the progression of cerebral ischemia/reperfusion injury via miR-126/SOX6 signaling pathway [J]. Biochem Biophys Res Commun. 2019. Jun 25;514(2):550–557.
- Wei R, Zhang L, Hu W, et al. Long non-coding RNA AK038897 aggravates cerebral ischemia/reperfusion injury via acting as a ceRNA for miR-26a-5p to target DAPK1 [J]. Exp Neurol. 2019. Apr;314:100-110. doi: 10.1016/j.expneurol.
- Zhong Y, Yu C, Qin W. LncRNA SNHG14 promotes inflammatory response induced by cerebral ischemia/reperfusion injury through regulating miR-136-5p/ROCK1 [J]. Cancer Gene Therapy. 2019. Jul;26(7-8):234–247.
- Liang X, Lin L, Woodling NS, et al. Signaling via the prostaglandin E2 receptor EP4 exerts neuronal and vascular protection in a mouse model of cerebral ischemia [J]. J Clin Invest. 2011;121(11):4362–4371.
- Zhang J, Yuan L, Zhang X, et al. Altered long non-coding RNA transcriptomic profiles in brain microvascular endothelium after cerebral ischemia [J]. Exp Neurol. 2016;277:162–170.
- Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs [J]. Cell. 2009;136(4):629–641.
- Ballantyne MD, Pinel K, Dakin R, et al. Smooth Muscle Enriched Long Non-Coding RNA (SMILR) regulates cell proliferation [J]. Circulation. 2016;133(21):2050.
- Samimi H, Haghpanah V, Irani S, et al. Transcript-level regulation of MALAT1-mediated cell cycle and apoptosis genes using dual MEK/Aurora kinase inhibitor “BI-847325” on anaplastic thyroid carcinoma [J]. DARU J Pharm Sci. 2019;27(1):1.
- Li J, Wang J, Chen Y, et al. LncRNA MALAT1 exerts oncogenic functions in lung adenocarcinoma by targeting miR-204 [J]. Am J Cancer Res. 2015;6(5):1099.
- Hochberg-Laufer H, Schwed-Gross A, Neugebauer KM, et al. Uncoupling of nucleo-cytoplasmic RNA export and localization during stress [J]. Nucleic Acids Res. 2019.May 21;47(9):4778–4797.
- Jia X, Shi L, Wang X, et al. KLF5 regulated lncRNA RP1 promotes the growth and metastasis of breast cancer via repressing p27kip1 translation [J]. Cell Death Dis. 2019;10(5):373.
- Li J, Chen C, Ma X, et al. Long noncoding RNA NRON contributes to HIV-1 latency by specifically inducing tat protein degradation [J]. Nat Commun. 2016;7:11730.
- Ding X, Jia X, Wang C, et al. A DHX9-lncRNA-MDM2 interaction regulates cell invasion and angiogenesis of cervical cancer [J]. Cell Death Differ. 2019 Sep;26(9):1750–1765
- Zhao J, Du P, Cui P, et al. LncRNA PVT1 promotes angiogenesis via activating the STAT3/VEGFA axis in gastric cancer [J]. Oncogene. 2018;37(30):4094.