Abstract
It was investigated that TP73-AS1(TP73 antisense RNA 1) could function as an oncogene in gastric cancer (GC). The expression and function of long noncoding RNAs (lncRNAs) could be impacted by single nucleotide polymorphisms (SNPs), which are related to cancer susceptibility and prognosis. This study was to reveal the association between lncRNAs TP73-AS1 polymorphisms (rs1181865 A > G, rs9800 G > C, rs3737589 A > G, rs2298222 G > A, rs7515164 C > A) and GC in 1000 GC cases and 1000 controls in a Chinese Han population. Rs3737589 G allele had significant associations with the increasing risk of GC (G vs. A: p = .005). Rs3737589 variant genotypes (AG + GG) were related to an increased risk of GC in the elder population (age ≥60), females, nonsmokers, nondrinkers, individuals living in urban, and individuals without family history of GC in stratified analyses. Rs3737589 variant genotypes (AG + GG) were related to the advanced depth of tumor invasion (T3 + T4). Besides, we found that GC patients with AG or GG genotype of rs3737589 had poorer overall survival (OS) than those with AA genotype (p < .05). Our findings showed that the lncRNA TP73-AS1 rs3737589 polymorphism might increase the risk of GC, and rs3737589 polymorphism could be a potential biomarker to predict the prognosis of GC patients.
Introduction
Gastric Cancer (GC) is one of the most common cancers in the world, being the fifth most common cancer and the third leading cause of cancer death worldwide [Citation1]. In China, the data from the National Central Cancer Registry of China (NCCR) shows that GC ranks second in terms of incidence and mortality among all cancer [Citation2]. The genesis and development of GC are influenced by many risk factors, including genetic risk factors [Citation3]. Increasing evidence identify that genetic polymorphisms are related to the risk and survival of GC [Citation4–6]. Indeed, the interrelation between single nucleotide polymorphisms (SNP) of genes and GC susceptibility has been recognized in our previous studies [Citation7–10].
Long noncoding RNAs (lncRNAs) are longer than 200 nucleotides noncoding RNAs. lncRNAs do not perform transcriptional tasks, but they can affect gene expression at the epigenetic, transcriptional and post-transcriptional levels [Citation11]. Emerging evidence recognizes that lncRNAs play imperative roles in various diseases, including cancer [Citation12]. TP73-AS1, a lncRNA transcribed from chromosome 1p36, has been regarded as a regulator in various cancers, such as gastric cancer [Citation13], breast cancer [Citation14], bladder cancer [Citation15], colorectal cancer [Citation16], clear cell renal cell carcinoma [Citation17], hepatocellular carcinoma [Citation18] and brain glioma [Citation19]. A previous study [Citation20] revealed that lncRNA TP73-AS1 was highly expressed in GC and lncRNA TP73-AS1 could promote cancer cell proliferation and invasion. Besides, they also found that the level of TP73-AS1 expression was positively related to lymph node metastasis, TNM stage, poor disease-free survival (DFS) and overall survival (OS) rates. Ding, Z et al. [Citation13] indicated that lncRNA TP73-AS1 could play the role of a competing endogenous RNAs (ceRNA) to bind to the microRNA (miR) miR-194-5p and improve the SDA1 domain containing 1 (SDAD1) expression to accelerate the genesis of GC.
Considering the tumorigenic function of TP73-AS1 in GC, we carried out this study to explore the association between TP73-AS1 polymorphisms (rs1181865, rs9800, rs3737589, rs2298222, rs7515164) and the risk and prognosis of GC in a Chinese Han population.
Materials and methods
Cases and controls
This case-control study randomly enrolled 1000 GC patients and 1000 cancer-free controls, who got medical treatment in the First Affiliated Hospital of Nanjing Medical University from 2012 to 2018. The study participants were Han Chinese who lived in Nanjing or surrounding cities. The patients without a history of other cancer in the case were diagnosed GC depending on pathological examination results. By observing the changes in cell structure and morphology of tissue slices with HE staining under light microscope, at least two pathologists determined together whether the tissue was gastric cancer. All controls did not have a history of any malignancies and gastric polyp, who were mainly from vascular surgery or orthopedics department and got medical treatments for some benign diseases. All subjects did not receive blood transfusion in the preceding six months. We gathered relevant information such as age, sex, hypertension, diabetes, smoking and drinking habits, living residence, family history (FH) and clinical data (cancer differentiation, depth of cancer infiltration, lymph node metastasis, tumor site) depended on questionnaires or medical records. our previous studies [Citation8] had defined hypertension, diabetes, smoker, and drinker. In the case, a total of 369 patients who accepted surgical treatment between January 2012 and May 2017 were followed up through telephone conversations at regular intervals. We defined survival time in the study was from the date of surgery to the date of follow-up or death, and the last follow-up date was April 2019. This study was approved by the ethics committee of the First Affiliated Hospital of Nanjing Medical University. Informed consent forms were obtained from each subject.
SNP selection
We selected tag SNPs of TP73-AS1 in chromosome 1, position 3735511–3747373 basing on 1000 Genomes Project (Ensembl GRCh38.p12). Using Haploview version 4.2 software (Cambridge, MA, USA) to choose the tag SNPs, five eligible tag SNPs were determined in the present research: rs1181865, rs9800, rs3737589, rs2298222, rs7515164. More details could be found in our previous studies [Citation9].
DNA extraction and genotyping
We obtained 5 ml venous blood from each subject and extracted DNA by a standardized kit following the manufacturer’s protocols as presented previously [Citation21]. The selected SNPs (rs1181865, rs9800, rs3737589, rs2298222, rs7515164) were genotyped by the polymerase chain reaction-ligase detection reaction (PCR-LDR) sequencing method. The length of DNA fragments and the corresponding primer sequences can be seen in Supplemental Table S1. PCR primer sequences were designed by using Primer-BLAST (http://www.ncbi.nlm.nih.gov/tools/primer-blast). The multiplex PCR was carried out in the reaction volume of 15 µl containing 2 µl DNA template, 7.5 µl 2× HotTaq PCR Reaction Mix, 2 µl mixed primers, and 3.5 µl ddH2O. The reaction conditions were as follows: pre-denaturation at 94 °C for 3 min, followed by 35 cycles of 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 90 s, at last extension at 72 °C for 3 min. LDR was performed in the reaction volume of 10 µl containing 3 µl multiplex PCR product, 1 µl 10 × Taq DNA ligase buffer, 0.125 µl 40 U/μl Taq DNA ligase, and 3 µl 1 pmol probes (0.2 µl each of probe), supplemented with ddH2O to reach the total volume of 10 µl. The corresponding probe sequences can be seen in Supplemental Table S2. Probe sequences are fixed sequences before and after the SNP sites in TP73-AS1. The reaction conditions were 30 cycles of 94 °C for 30 s and 56 °C for 3 min. The LDR products were analyzed on an ABI 3730XL DNA sequencer. This work was carried out by the Shanghai Generay Biotech Co. Ltd (http://www.generay.com.cn). In addition, 10% of the samples were selected randomly to repeated assays for quality control, and the final concordance rate was 100%.
Real-time PCR analyses of TP73-AS1 expression levels
Eighty GC tissues with 40 AA, 35 AG and 5 GG genotype of rs3737589 were stored with liquid nitrogen immediately and extracted total RNA through TRIzol reagent (Invitrogen, Carlsbad, CA, USA). Followed by the protocol of the manufacturer, we used Primescript RT Reagent (Takara, Japan) to reverse transcribe the total RNA into cDNA. Finally, relative TP73-AS1 expression was evaluated by quantitative real-time PCR with a StepOnePlus RealTime PCR System (Applied Biosystems) by using the SYBR way. Primer sequences of TP73-AS1 were: forward: 5′-CCGGTTTTCCAGTTCTTGCAC-3′; reverse: 5′-GCCTCACAGGGAAACTTCATGC-3′. The GAPDH served as the endogenous control to standardize data. Its primer sequences were: forward 5’-TCCTCTCCCAAGTCCACACA-3′; reverse 5’-GCACGAAGGCTCATCATTCA-3′.
Statistical analysis
SPSS (Version 22.0, IBM, Armonk, NY, USA) was performed to analyze the data. Student’s t-tests and χ2 tests were applied to identify differences in demographic variables between the case and control groups. The goodness-of-χ2 test was used to analyze the Hardy-Weinberg equilibrium among control subjects. The relations of alleles/genotypes with GC susceptibility were evaluated by odds ratio (OR) and 95% confidence interval (CI) by using the methods of the unconditional univariate analysis. Adopting 2−ΔCT algorithm, we calculated the relative expression levels of TP73-AS1 in 80 GC tissues. ANOVA was applied to identify the relations of the TP73-AS1 polymorphism with the expression levels of TP73-AS1. Survival curves were calculated by the Kaplan–Meier method, and differences between curves were analyzed by the log-rank test. Mean survival time (MST) was provided because the median survival time could not be figured out. The relation between the SNP and the OS of GC patients was estimated by calculating the hazard ratio (HR) with 95%CI in univariate analyses and adjusted HR with 95%CI in multivariate analyses. P values <.05 was considered to be statistically significant.
Results
Characteristics of the subjects
The basic information of 1000 cases and 1000 controls were listed in . The distribution of age, sex, hypertension, and diabetes was not statistically different between the two groups (p > .05). However, the ratio of smokers (28.2% vs. 19.3%, p < .001) and drinkers (21.1% vs. 13.6%, p < .001) in cases was remarkably higher than the ratio in controls. Meanwhile, the differences between the GC patients and cancer-free controls in terms of residence (p < .001) and family history (p < .001) are significant. The proportion of patients who lived in rural and had a family history in the GC group was significantly higher than that in controls.
Table 1. Demographic information.
Associations of SNPs in TP73-AS1 with GC risk
The genotype and allele frequencies of TP73-AS1 tag SNPs and their relations with GC risk in a series of models were displayed in . In controls, genotype frequencies of the five SNPs all met the Hardy-Weinberg equilibrium (p > .05). For rs3737589, the ratio of G allele in the cases was statistically higher than the controls (p = .005). Taking AA genotype as a comparison, AG, GG and AG + GG genotypes were significantly associated with increased risk of GC (p = .021, .016, .006 and .005 respectively). Rs1181865 G allele was also related to a more remarkably increased risk of GC than the C allele. (p = .010, OR = 1.18, 95% CI = 1.04– 1.33). However, there was no statistical significance between the three SNPs (rs9800, rs2298222, rs7515164) and GC risk (p > .05, Supplemental Table S3).
Table 2. Association between TP73-AS1 gene polymorphisms and risk of gastric cancer (rs1181865, rs3737589).
Stratified analysis for the polymorphism and risk
To exclude the potential confounders which may affect the association of the polymorphism with GC risk, stratified analysis for rs1181865 and rs3737589 was adopted. The results are shown in and . For rs1181865, the increased risk connected with the AA + AG genotypes are more evident in non-drinkers (adjusted OR = 1.32, 95% CI = 1.05– 1.65; p = .016), and individuals with family history (p = .019 and .042 respectively) but not in drinker (adjusted OR = 0.89, 95% CI = 0.54– 1.46; p = .642) and individuals without family history (adjusted OR = 0.62, 95% CI = 0.11– 3.45; p = .586). besides, the association between the polymorphism and GC risk in terms of age, sex, smoking status, and the living place was not statistically significant.
Table 3a. Stratified analyses for TP73-AS1 genotypes in cases and controls (rs1181865).
Table 3b. Stratified analyses for TP73-AS1 genotypes in cases and controls (rs3737589).
For rs3737589, We detected an obviously increased susceptibility of GC in age ≥60 years subjects (p = .009, adjusted OR = 1.37, 95% CI = 1.08–1.72), females (p = .001, adjusted OR = 1.71, 95% CI = 1.25–2.34), nonsmokers (p = .005, adjusted OR = 1.35, 95% CI = 1.10–1.65), nondrinkers (p = .002, adjusted OR = 1.37, 95% CI = 1.13–1.67), and individuals living in urban areas (p = .033, adjusted OR = 1.34, 95% CI = 1.03–1.76) and individuals without family history (p = .005, adjusted OR = 1.30, 95% CI = 1.08–1.56). Nevertheless, the association was not found in the opposite subgroups. The interaction between the genotypes and clinicopathologic characteristics of GC also was summed up in and Supplemental Table S4. We found that GC patients with rs3737589 AG or GG genotype could be likely to suffer the more serious depth of infiltration (T3 + T4) compared with rs737589 AA genotype (adjusted HR = 1.36, 95%=1.04–1.77, p = .023). However, the association for rs1181865 was not discovered.
Table 4. Associations between variant TP73-AS1 genotypes and clinicopathologic characteristics of gastric cancer (rs3737589).
In silico prediction of effects of SNPs on TP73-AS1
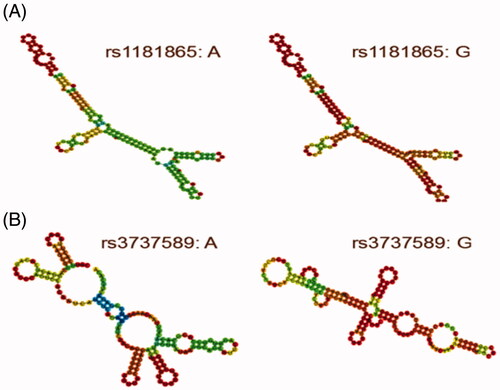
Considering that rs1181865 (A > G) and rs3737589 (A > G) are located in the exon region of TP73-AS1 gene which means the SNPs could be completely reserved throughout the process of gene expression, we analyzed the impact of rs1181865 and rs3737589 on the local secondary structure of TP73-AS1 by performing the RNAfold algorithm [Citation22] in silico. The local folding structure of TP73-AS1 was slightly different depending on the allele of rs1181865 changes but was apparently affected by the changes of the rs3737589 allele in . Furthermore, using the prediction function of the RegRNA2.0 program [Citation23], we found that rs1181865 (A > G) might contribute to the loss of hsa-miR-3189-5p, hsa-miR-328-3p and hsa-miR-141-5p binding site or the gain of hsa-miR-7162-5p, hsa-miR-516b-3p binding site and rs3737589 (A > G) might contribute to the gain hsa-miR-7848-3p binding site on TP73-AS1.
Functional relevance of the SNPs to TP73-AS1 expression
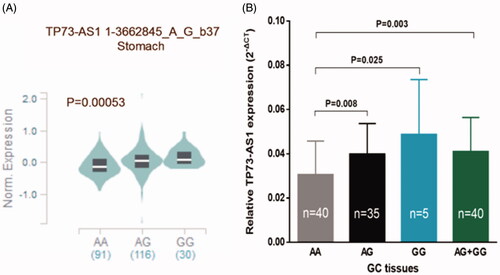
Depending on a public database, GTEx portal (https://gtexportal.org/home/), we further explored whether rs1181865 and rs3737589 could affect the TP73-AS1 expression level in gastric tissues. We only got the data about rs3737589 (), people who carried the AG/GG genotypes had apparently higher TP73-AS1 expression levels in normal stomach tissues than those carrying AA genotype (p < .005). Similarly, GC tissues carrying AG, GG or AG + GG genotypes of rs3737589 had a significantly higher TP73-AS1 expression level (mean ± standard error) when compared with those carrying AA genotype (p = .008, .025, and .003, resp.).
Figure 2. The effect of rs3737589 on TP73-AS1 expression level. (A): The genotype of rs3737589 and expression of TP73-AS1 gene in stomach was based on the public GTEx portal database. (B): TP73-AS1 levels were determined by qRT-PCR in GC tissues in subjects with the AA (n = 40), AG (n = 35) or GG (n = 5) genotype of rs3737589. Results were shown as mean ± standard error relative to GAPDH levels.
Association of the GC patients’ clinicopathologic features and overall survival with rs3737589
Just as shown, we found that the survival time had significant associations with the depth of invasion and lymph node metastasis (p < .001), while we did not discover the associations in the term of age, sex and tumor sites. These conclusions above were accordant in Cox regression analysis (). Considering the significant association between rs3737589 AG + GG genotype and the advanced depth of tumor invasion (T3 + T4), we assessed the relationship between genotypes of rs3737589 and the OS of GC patients which was summed up in . Kaplan Meier analysis indicated that GC patients carrying AG/GG genotype had a poorer OS than those carrying AA genotype (in , AG vs. AA, log-rank p = .030; GG vs. AA, log-rank p = .006; AG + GG vs. AA log-rank p = .013). In further univariate and multivariate Cox regression analysis, we confirmed that rs3737589 is an independent risk factor in OS of GC patients.
Figure 3. Kaplan-Meier survival curves for the overall survival by characteristics and clinical features of gastric cancer patients. (age, sex, sites, depth of invasion, lymph nodes metastasis.).
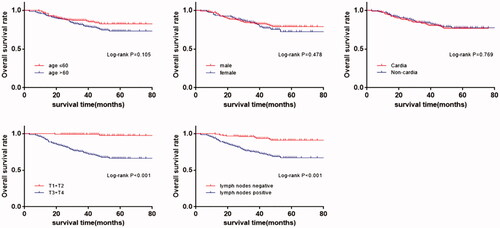
Figure 4. Kaplan-Meier survival curves of rs3737589 polymorphism for the overall survival in patients with gastric cancer. (AA vs. AG, AA vs. GG, AA vs. AG + GG.).
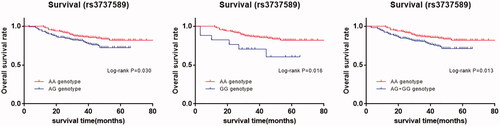
Table 5. Survival analysis by clinicopathologic features of gastric cancer patients.
Table 6. Association between genotype of rs3737589 and overall survival in gastric cancer patients.
Discussion
In the study, we discovered that rs1181865 (AG/GG) and rs3737589 (AG/GG) genotypes were obviously related to increased risk of GC, and genotypes of rs3737589 (AG/GG) could obviously decrease the OS of GC patients who accepted surgery treatment. Furthermore, TP73-AS1 expression levels in cancer tissues were varied distinctly according to rs3737589 genotypes, and the result is consistent with the GTEx portal in normal tissues. The roles of lncRNA have attracted extensive attention, especially in the initiation and progression of tumors [Citation24]. TP73-AS1 had been reported to be over-expressed in gastric cancer, which not only promotes tumor occurrence and development, but also could predicts unfavorable prognosis for gastric cancer [Citation25]. Meanwhile, depending on the advent of genome wide association studies (GWAS) and high-throughput genetic analysis, many researches demonstrated particular SNPs can alter the gene expression profile and affect gene function and finally lead to increased risk of susceptibility to many diseases, including cancer [Citation26]. In this study, we identified that the level of TP73-AS1 with rs3737589 G is higher than with rs3737589 A in normal gastric tissues or GC tissues. However, the exact mechanisms for rs3737589 how to decrease the expression level of TP73-AS1 is still not understood. Some studies had pointed out that SNPs can influence the RNA stability which led to the change of the RNA function [Citation27]. By using the RNAfold algorithm, we indeed predicted that the change of the folding structure by rs3737589 was more obvious. Basing on the prediction, we speculated that the changes of the local folding structure may lead some locus exposure or hide which can impact the interaction with other molecules, and at last result in the change of gene expression. Accumulating evidence has verified that miRNA participated in various malignant tumors and some lncRNAs could play the role of ceRNA to sponge related miRNAs to regulate the expression level of other transcripts [Citation28]. By the prediction tool of RegRNA2.0, we indeed found rs1181865 and rs3737589 polymorphisms could lead the gains and losses of miRNA binding sites for TP73-AS1. So, we speculated that theTP73-AS1 genetic variants may change target miRNAs and eventually affect the function of TP73-AS1. However, all the hypotheses about specific mechanisms need to be verified by further investigations in the future.
In the stratified analysis, we revealed that rs3737589 polymorphism was obviously related to increased GC risk in the subjects of older individuals (aged ≥60), women, non-smokers, no-drinkers, rural areas and individuals without FH, but without significance in the opposite subgroups. These results may be explained in the following inferences. Initiation and progression of gastric cancer contribute to complex genetic and environmental interactions [Citation29].
The older individuals (age ≥60) who carry the rs3737589 AG + GG genotypes were more likely to suffer from GC, which meant that the risk effects of these genotypes could be more highlighted in the older people. Smoking and drinking have been considered as independent risk factors in the occurrence and development of gastric cancer. These strong risk factors may dissimulate the relation of rs3737589 and GC risk and ultimately contribute that the association is more significant in terms of non-smokers and non-drinkers. Similarly, individuals living in rural areas may be more likely to form a relatively unhealthy diet habit, such as pickled food. High-salt diet has been recognized as a strong risk factor interacting with helicobacter pylori infection for the development of GC [Citation30]. Patients with FH might carry a series of stronger risk genetic factors, which lead to the effect of rs3737589 variant genotypes for GC risk in these patients may be submerged. The explanation for the FH subgroup might be similar to the smoker subgroup and the drinker subgroup. GC is more common in males (twice as common as in women). It means the male may have more risk factors than the female which submerge the risk of rs3737589 similarly. Hence, the interaction of rs3737589 and GC risk is more obvious in the female. However, these findings and the mechanism should be validated by further studies with a larger-scale population in the future.
Several limitations in the case-control study should not be neglected. Firstly, some other environmental factors are also risk factors for gastric cancer, such as high-salt diet, chronic gastric ulcer, and high body weight, especially helicobacter pylori infection. Besides, in terms of prognosis, we failed to get the information that whether these patients after accepting surgery treatment got other treatments like chemotherapy, radiotherapy, and biotherapy which may have an unforeseen influence on overall survival in GC patients. Secondly, the relatively deficient sample size and the selective bias of subjects should not be ignored. Finally, our study was based on a Chinese Han population, the results may be not applicable to other regions and ethnic groups.
Conclusion
In short, we uncovered that rs2288947 and rs3737589 in TP73-AS1 were significantly related to GC risk, and rs3737589 was obviously related to a poorer OS of GC patients in a Chinese population. These findings showed that the polymorphisms of TP73-AS1 could assess susceptibility to GC in population. Furthermore, rs3737589 polymorphism may be a useful potential biomarker for the prognosis of GC patients.
Supplementary_Materials_2.1_.docx
Download ()Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018; 68(6):394–424.
- Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132.
- Karimi P, Islami F, Anandasabapathy S, et al. Gastric cancer: descriptive epidemiology, risk factors, screening, and prevention. Cancer Epidemiol Biomarkers Prev. 2014;23(5):700–713.
- He B, Pan B, Pan Y, et al. Polymorphisms of IL-23R predict survival of gastric cancer patients in a Chinese population. Cytokine. 2019; 117:79–83.
- Wang W, Li Z, Wang J, et al. A functional polymorphism in TFF1 promoter is associated with the risk and prognosis of gastric cancer. Int J Cancer. 2018;142(9):1805–1816.
- Gu H, Yang L, Tang N, et al. Association of endothelin-converting enzyme-1b C-338A polymorphism with gastric cancer risk: a case-control study. Eur J Cancer. 2008;44(9):1253–1258.
- Huang C, Wang Y, Fan H, et al. Association analysis of DACT1 genetic variants and gastric cancer risk in a Chinese Han population: a case-control study. OTT. 2016;9:5975–5983.
- Ma X, Yang C, Tang R, et al. Association between LMP2 and LMP7 gene polymorphisms and the risk of gastric cancer: a case-control study. Oncol Lett. 2015;10(1):509–517.
- Ge Y, He Y, Jiang M, et al. Polymorphisms in lncRNA PTENP1 and the risk of gastric cancer in a Chinese population. Dis Markers. 2017;2017:1.
- Yang C, Ma X, Liu D, et al. Promoter polymorphisms of miR-34b/c are associated with risk of gastric cancer in a Chinese population. Tumor Biol. 2014;35(12):12545–12554.
- Kung JTY, Colognori D, Lee JT. Long noncoding RNAs: past, present, and future. Genetics. 2013;193(3):651–669.
- Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29(4):452–463.
- Ding Z, Lan H, Xu R, et al. LncRNA TP73-AS1 accelerates tumor progression in gastric cancer through regulating miR-194-5p/SDAD1 axis. Pathol Res Pract. 2018;214(12):1993–1999.
- Zou Q, Zhou E, Xu F, et al. A TP73-AS1/miR-200a/ZEB1 regulating loop promotes breast cancer cell invasion and migration. J Cell Biochem. 2018;119(2):2189–2199.
- Tuo Z, Zhang J, Xue W. LncRNA TP73-AS1 predicts the prognosis of bladder cancer patients and functions as a suppressor for bladder cancer by EMT pathway. Biochem Biophys Res Commun. 2018;499(4):875–881.
- Cai Y, Yan P, Zhang G, et al. Long non-coding RNA TP73-AS1 sponges miR-194 to promote colorectal cancer cell proliferation, migration and invasion via up-regulating TGFalpha. CBM. 2018;23(1):145–156.
- Liu G, Zhao X, Zhou J, et al. LncRNA TP73-AS1 promotes cell proliferation and inhibits cell apoptosis in clear cell renal cell carcinoma through repressing KISS1 expression and inactivation of PI3K/Akt/mTOR signaling pathway. Cell Physiol Biochem. 2018;48(1):371–384.
- Li S, Huang Y, Huang Y, et al. The long non-coding RNA TP73-AS1 modulates HCC cell proliferation through miR-200a-dependent HMGB1/RAGE regulation. J Exp Clin Cancer Res. 2017;36(1):51.
- Xiao S, Wang R, Wu X, et al. The long noncoding RNA TP73-AS1 interacted with miR-124 to modulate glioma growth by targeting inhibitor of apoptosis-stimulating protein of p53. DNA Cell Biol. 2018;37(2):117–125.
- Wang Y, Xiao S, Wang B, et al. Knockdown of lncRNA TP73-AS1 inhibits gastric cancer cell proliferation and invasion via the WNT/beta-catenin signaling pathway. Oncol Lett. 2018;16(3):3248–3254.
- Yang L, Gu HJ, Zhu HJ, et al. Tissue inhibitor of metalloproteinase-2 G-418C polymorphism is associated with an increased risk of gastric cancer in a Chinese population. Eur J Surg Oncol. 2008;34(6):636–641.
- Gruber AR, Lorenz R, Bernhart SH, et al. The Vienna RNA websuite. Nucleic Acids Res. 2008;36(Web Server):W70–4.
- Chang TH, Huang HY, Hsu JB, et al. An enhanced computational platform for investigating the roles of regulatory RNA and for identifying functional RNA motifs. BMC Bioinformatics. 2013;14(Suppl 2):S4.
- Evans JR, Feng FY, Chinnaiyan AM. The bright side of dark matter: lncRNAs in cancer. J Clin Invest. 2016;126(8):2775–2782.
- Zhang W, Zhai Y, Wang W, et al. Enhanced expression of lncRNA TP73-AS1 predicts unfavorable prognosis for gastric cancer and promotes cell migration and invasion by induction of EMT. Gene. 2018;678:377–383.
- McLean MH, El-Omar EM. Genetics of gastric cancer. Nat Rev Gastroenterol Hepatol. 2014;11(11):664–674.
- Lv Z, Xu Q, Sun L, et al. Four novel polymorphisms in long non-coding RNA HOTTIP are associated with the risk and prognosis of colorectal cancer. Biosci Rep. 2019;39(5):1–12
- Song YX, Sun JX, Zhao JH, et al. Non-coding RNAs participate in the regulatory network of CLDN4 via ceRNA mediated miRNA evasion. Nat Commun. 2017;8(1):289.
- Ashktorab H, Kupfer SS, Brim H, et al. Racial disparity in gastrointestinal cancer risk. Gastroenterology. 2017;153(4):910–923.
- Cheng XJ, Lin JC, Tu SP. Etiology and prevention of gastric cancer. Gastrointest Tumors. 2016;3(1):25–36.