Abstract
LINC00473 has been reported to be aberrantly expressed in diverse kinds of human malignancy. However, the function and underlying mechanisms of LINC00473 in glioma still remain unclear. In the present study, LINC00473 was notably elevated in glioma tissues and cell lines. High LINC00473 expression was associated with advanced WHO grade (III–IV), low Karnofsky performance score (KPS), and poor prognosis. Loss function assays showed that LINC00473 knockdown decreased glioma cell proliferation, invasion and epithelial–mesenchymal transition (EMT) processes in vitro, and reduced tumor growth in vivo. Mechanistic analysis indicated that LINC00473 regulated CDK6 expression by competitively binding to miR-637. Moreover, rescue assays revealed that miR-637 inhibitors abolished the effects of LINC00473 suppression on glioma cells progression. Thus, we demonstrated that LINC00473 could act as a ceRNA of miR-637 to promote glioma progression through regulating CDK6 expression, which provided a potential therapeutic target for treatment of glioma patients.
Introduction
Glioma is the most frequent intracranial malignant brain tumors in adults, characterized by rapid growth, early metastasis, and high lethality [Citation1,Citation2]. Despite accumulating achievements in treatments of glioma, the prognosis is still unsatisfactory [Citation3]. Although many studies have been performed to detect the molecules associated with gliomas, only a few have been revealed [Citation4,Citation5]. Therefore, it is urgent to discover novel molecular targets for therapy of glioma patients.
Long non-coding RNAs (lncRNAs) consist of a group of RNA molecules that are longer than 200 nucleotides in length with limited protein-coding potential [Citation6]. Increasing evidence showed that lncRNAs might play important roles in the regulation of gene transcription, translation and widespread regulators [Citation7–9]. LncRNA dysregulation contributed to the progression of multiple cancers. For example, Zhang et al. showed that high MALAT1 expression was correlated with advanced tumor progression and poor prognosis in clear cell renal cell carcinoma [Citation10]. Liu et al. found that lncRNA GHET1 promoted esophageal squamous cell carcinoma cells proliferation and invasion via EMT processes [Citation11]. Zhang et al. reported that lncRNA HNF1A-AS1 promoted lung cancer cell proliferation and invasion via regulating miR-17-5p expression [Citation12].
LINC00473 is an intergenic long noncoding RNA which has been reported to be aberrantly expressed in several malignancies. For example, Chen et al. showed that LINC00473 could act as a therapeutic target for LKB1-inactivated lung cancer [Citation13]. Zhu et al. found that LINC00473 acted as an oncogene to promote Wilms tumour growth via miR-195/IKKα axis [Citation14]. Shi et al. suggested that LINC00473 promoted cervical cancer progression by the miR-34a/ILF2pathway [Citation15]. However, the expression and biological functions of LINC00473 in glioma remain poorly understood.
In the present study, LINC00473 expression was significantly increased in glioma tissues and cell lines. High LINC00473 expression was associated with advanced clinical features and poor prognosis. In mechanism, we found that LINC00473 promoted glioma cells proliferation, invasion and EMT processes by acting as a ceRNA of miR-637 to regulate CDK6 expression. Taken together, we suggested that LINC00473/miR-637/CDK6 axis might act as a potential therapeutic target for glioma treatment.
Materials and methods
Tissue samples
A total of 31 glioma specimens and 14 normal brain tissues (NBTs) were obtained from The First Affiliated Hospital of Zhengzhou University and The First Affiliated Hospital, and College of Clinical Medicine of Henan University of Science and Technology. Diagnosis was based on pathological evidence, and collected tissue samples were immediately snap-frozen in liquid nitrogen and stored at −80 °C until used. All patients were not received preoperative treatments before surgery. Written informed consents were obtained from all participants. The study was approved by the Ethics Committee of The First Affiliated Hospital of Zhengzhou University and The First Affiliated Hospital, and College of Clinical Medicine of Henan University of Science and Technology.
Cell culture
Human glioma cell lines (LN229, LN18, LN308, U87, U251 and SNB19) and normal human astrocytes (NHA) were purchased from Chinese Academy of Medical Sciences (Beijing, China). All cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) medium with 10% fetal bovine serum (Gibco, NY, USA) supplemented with 100 units/mL penicillin and 100 mg/mL streptomycin in a humidified atmosphere containing 5% CO2 at 37 °C.
RNA extraction and qRT-PCR analyses
Total RNA was extracted from tissue samples or cultured cells with TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocols. RNA was reversely transcribed into cDNAs by using a Reverse Transcription Kit of the Prime-ScriptTM one step (Takara, Dalian, China). qRT-PCR was performed using the ABI7500 System (Applied Biosystems, CA, USA) and the SYBR Green PCR Master Mix kit (TaKaRa). After normalizing to the endogenous control GAPDH or U6, the relative expression was calculated utilizing the 2−△△Ct method. Each experiment was repeated in triplicate at least.
Western blot
Total protein from cells was extracted using RIPA assay and loaded for electrophoresis (Beyotime, Shanghai, China). After transferring on a PVDF membrane at 300 mA for 100 min, the membrane was blocked in 5% skim milk for 2 h, incubated with primary antibodies at 4 °C overnight. The membranes were then treated with horseradish peroxidase-conjugated secondary antibodies for 2 h. The bands were detected using an ECL Detection Systems (Thermo Scientific, Beijing, China).
MTT assay
MTT assay was used to detect cell proliferation. Briefly, transfected cells were seeded in 96‐well plate (2000 cells per well) and culture for 24 h, 48 h, 72 or 96 h. Then transfected cells were incubated with 20 μL (10 mg/mL) MTT solution was added into each well and incubated at 37 °C for another 4h. The absorbance was measured at a wavelength of 490 nm.
Colony formation assay
For colony formation assay, transfected cells were plated into 6-well plates and then incubated at 37 °C for 2 weeks. The colonies were subsequently washed with PBS, fixed with methanol and then stained with crystal violet. A coulter counter was then utilized to count the cell colonies.
Transwell invasion assay
Transwell invasion assay was processed according to manufacturer’s instructions and previous study [Citation10].
Luciferase reporter assay
The wild-type (WT) and mutated-type (MUT) 3’UTR with LINC00473 were synthesized by site mutation method, amplified by PCR and then were inserted into pmirGLO (Promega, Madison, Wisconsin, USA). Afterwards the Wt-LINC00473 or Mut-LINC00473 constructs were transfected into cells with the company of miR-637 mimics or miR-NC, the process was implemented inside a 24-well plates. After 48 h, luciferase activity was detected using the Dual-Luciferase Reporter Assay system (Promega). Renilla luciferase activities were used for normalization.
RIP assay
RIP assay was conducted on cells by using EZ-Magna RIP Kit (Millipore, Billerica, MA, USA) following the manufacturer’s instructions. The cells were lysed in complete RNA lysis buffer, then incubated using RIP buffer that contains magnetic beads conjugated with human antibody (Millipore) or negative control mouse IgG. The protein was digested using Proteinase K, and immunoprecipitated RNA was isolated. Purified RNA was subjected to qRT-PCR analysis.
In vivo tumorigenesis assays
Nude athymic BALB/c mice (6-week-old) were purchased from the Laboratory Animal Center of Southern Medical University, and all procedures were approved by the Ethics Committee of the Fifth Affiliated Hospital of Zhengzhou University. For tumorigenesis assay, the U251 cells transfected with sh-NC or sh-LINC00473 were injected subcutaneously into the flank of nude mice, followed by measurements of the tumor volume at 7-day intervals using the formula: volume = 0.5 × length × width2. At 7 weeks after injection, the mice were sacrificed, and tumors were harvested for the determination of tumor mass as well as the IHC analysis of the expression levels of Ki-67.
Statistical analyses
The whole data analysis employed SPSS 17.0 software. The format of mean ± standard deviation was used to present the results. Two-tailed Student’s t-test, or One-way analysis of variance was applied for statistical analysis. p < .05 was recognized to have statistical significant difference.
Results
LINC00473 is upregulated in glioma
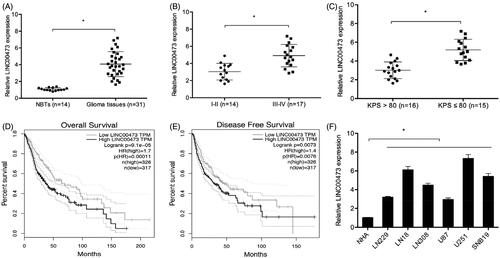
In the present study, qRT-PCR was used to detect LINC00473 expression in 31 paired glioma tissues and normal brain tissues (NBTs). QRT-PCR showed that LINC00473 expression was increased in glioma tissues (. High LINC00473 expression was associated with advanced WHO grade, and low KPS score (. Patients displaying a high expression of LINC00473 contributed to poor overall survival (OS) and disease free survival (DFS) (. Moreover, LINC00473 expression was significantly upregulated in glioma cells (LN229, LN18, LN308, U87, U251 and SNB19) compared to NHA cells (Figure1(F)). These data suggested that LINC00473 might play critical roles in glioma progression.
Figure 1. LINC00473 was upregulated in glioma. (A) LINC00473 was highly expressed in glioma tissues than normal tissues. (B and C) LINC00473 was upregulated in glioma patients with advanced WHO grade (III–IV) and low KPS score. (D and E) High LINC00473 expression was associated with poor OS and DFS in glioma patients. (F) LINC00473 was highly expressed in glioma cell lines (LN229, LN18, LN308, U87, U251 and SNB19) compared to NHA cells. *p < .05.
LINC00473 promotes glioma proliferation and invasion
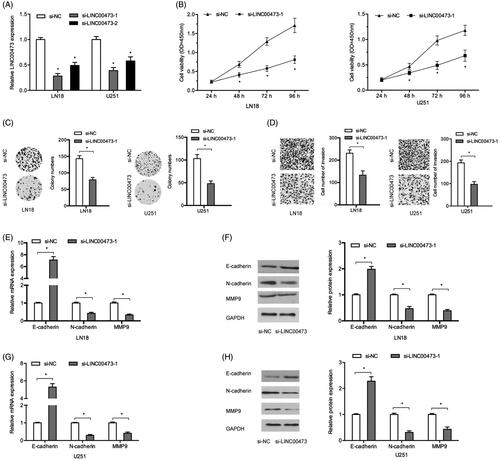
To investigate the roles of LINC00473 on tumor progression, glioma cells were transfected with si-LINC00473 or si-NC. QRT-PCR showed that si-LINC00473 significantly reduced LINC00473 expression in LN18 and U251 cells, especial in si-LINC0073-1 (. MTT and colony formation assays showed that LINC00473 suppression significantly decreased LN18 and U251 cells viability (. Transwell assay showed that LINC00473 inhibition reduced the invasion capacity of LN18 and U251 cells (. To further determine the underlying mechanism, qRT-PCR and Western blot assays were used to determine MMP9 (invasion marker), E-cadherin (the epithelial marker), and N-cadherin (the mesenchymal markers) expression in glioma cells. Results showed that LINC00473 suppression significantly reduced MMP9, N-cadherin expression, while increased E-cadherin expression both in mRNA and protein levels (.
Figure 2. LINC00473 suppression reduced glioma cells progression. (A) si-LINC00473 transfection efficiency detected by qRT-PCR in LN18 and U251 cells. (B and C) LINC00473 suppression reduced LN18 and U251 cells proliferation and colony numbers. (D) LINC00473 inhibition decreased the invasion capacity of LN18 and U251 cells. (E -H) MMP9, E-cadherin, and N-cadherin expression in LN18 and U251 cells transfected with si-LINC00473 or si-NC. *p < .05.
LINC00473 negatively regulates miR-637 in glioma cells
Increasing evidence showed that cytoplasmic lncRNAs could function as sinks for pools of active miRNAs, functionally liberating mRNAs to mediate several biological processes [Citation16]. Thus, we firstly explored the distribution of LINC00473 by lncLocator. Results showed that the vast majority of LINC00473 was located in the cytoplasm (), and the result was further confirmed by qRT-PCR (. Subsequently, DIANA was used to predict the target of LINC00473, and result represented that LINC00473 could target miR-637 (. Luciferase reporter assay revealed that miR-637 overexpression significantly decreased the luciferase activity of Wt-LINC00473 rather than Mut-LINC00473 (. RIP assay revealed that LINC00473 and miR-637 were both enriched in the immunoprecipitation (. QRT-PCR showed that si-LINC00473 significantly increased miR-637 expression in LN18 and U251 compared to si-NC group (.
Figure 3. LINC00473 acted as a sponge of miR-637 in glioma cells. (A and B) lncLocator and qRT-PCR showed that LINC00473 is located in the cytoplasm. (C) LINC00473 information and the binding sites between miR-637 and LINC00473. (D) miR-637 mimics reduced the luciferase activity of the Wt-LINC00473 group. (E) RIP assay revealed that LINC00473 and miR-637 expression were both enriched in the immunoprecipitation. (F) LINC00473 inhibition increased miR-637 expression in LN18 and U251 cells. (G) miR-637 was decreased in glioma tissues. (H and I) Low miR-637 expression was correlated with high WHO grade (III–IV) and low KPS score. (J) High LINC00473 expression was negatively correlated with miR-637 expression in glioma tissues. *p < .05.
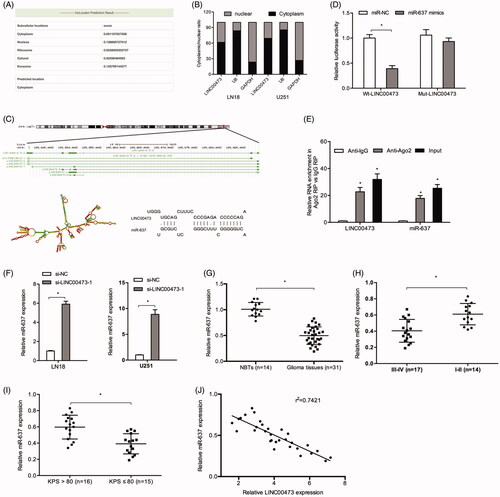
Next, the expression level of miR-637 was also measured in glioma tissues, results showed that miR-637 expression was decreased in glioma tissues (. Low miR-637 expression was associated with advanced WHO grade, and low KPS score (. Moreover, our results showed that the expression of miR-637 was negatively correlated with LINC00473 expression in glioma tissues (.
CDK6 is targeted by miR-637
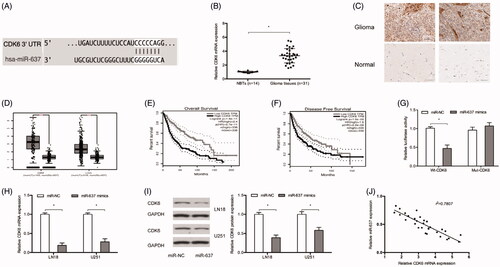
It has been shown that miRNAs could regulate gene expression by directly binding with the 3’UTR of their target genes [Citation17]. TargetScan showed that CDK6 have a potential binding sites of miR-637 (. Next, we explored CDK6 expression in glioma, results showed that CDK6 was significantly increased in glioma (), which was consistent with TCGA database results (. Kaplan Meier analysis showed that high CDK6 expression was associated with poor OS and DFS of patients with glioma (. Luciferase reporter assay suggested that miR‐637 mimics reduced the luciferase activity of Wt-CDK6 rather than Mut-CDK6 group (. Ectopic expression of miR-637 reduced CDK6 expression both in mRNA and protein levels in LN18 and U251 cells (. Moreover, CDK6 expression was negatively correlated with miR-637 expression in glioma tissues (.
Figure 4. CDK6 was targeted by miR-637. (A) Predicted binding sites between miR-637 and CDK6. (B and C) CDK6 was upregulated in glioma tissues both in mRNA and protein levels. (D) TCGA database showed that CDK6 expression was upregulated in glioma tissues. (E and F) Kaplan Meier analysis showed that high CDK6 expression was associated with poor OS and DFS of patients with glioma. (G) miR-637 mimics reduced the luciferase activity of the Wt-CDK6 group. (H and I) MiR-637 mimics reduced CDK6 expression both in mRNA and protein levels. (J) CDK6 expression was negatively correlated with miR-637 expression in glioma tissues. *p < .05.
LINC00473/miR-637/CDK6 axis promotes glioma progression
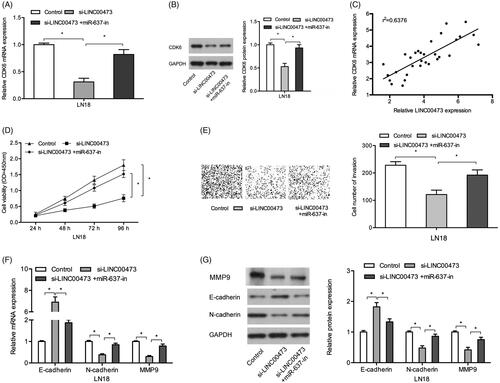
Next, we explored whether LINC00473 could act as a ceRNA in regulating CDK6 by competitively binding to miR-637 in glioma. QRT-PCR and Western blot showed that CDK6 expression was reduced by LINC00473 inhibition in LN18 cells, and the reduction could be reversed by miR-637 inhibitors (. Correlation analysis showed that LINC00473 expression was positively correlated with CDK6 expression in glioma tissues (. Moreover, rescue assays showed that the effects of LINC00473 inhibition on glioma cells proliferation and invasion could be abolished by miR-637 inhibitors (. Similarly, qRT-PCR and Western blot results showed that miR-637 inhibition could reverse the effects of LINC00473 inhibition on N-cadherin, MMP9, and E-cadherin expression in LN18 cells (. These data indicated that LINC00473 could promote glioma cells progression by regulating the miR-637/CDK6 axis.
Figure 5. LINC00473/miR-637/CDK6 axis in glioma progression. (A and B) The effects of LINC00473 suppression on CDK6 could be reversed by miR-637 inhibitors in LN18 cells. (C) LINC00473 expression was positively correlated with CDK6 expression in glioma tissues. (D and E) LINC00473 inhibition on LN18 cells proliferation and invasion could be rescued by miR-637 inhibitors. (F and G) The effects of LINC00473 inhibition on E-cadherin, N-cadherin and MMP9 expression could be rescued by miR-637 inhibitors in LN18 cells. *p < .05.
LINC00473 promotes tumor growth in vivo
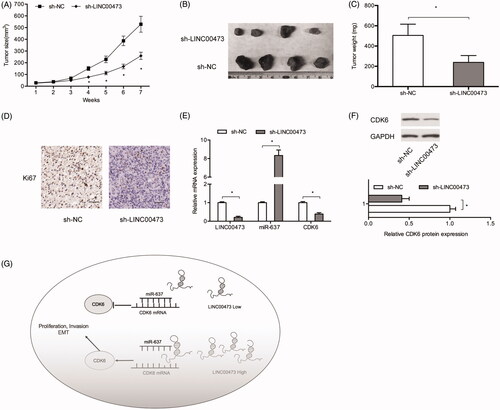
To explore the roles of LINC00473 in vivo, a xenograft mouse model was used. Results showed that tumor growth and weights were significantly suppressed in sh-LINC00473 group compared to sh-NC group (). Ki67 assay indicated that LINC00473 inhibition reduced tumor cell proliferation (. Moreover, we explored LINC00473/miR-637/CDK6 axis expression in xenograft tissues, results showed that LINC00473 expression and CDK6 expression were significantly decreased, while miR-637 expression was upregulated in sh-LINC0047 (. Thus, we suggested that LINC00473 could promote glioma progression by impairing the miR-637/CDK6 axis (.
Figure 6. LINC00473 inhibition reduced tumor growth in vivo. (A–C) LINC00473 downregulation reduced tumor growth and weights in vivo. (D) Ki67 expression was determined by IHC in xenograft tissues. (E) Relative expression of LINC00473, miR-637 and CDK6 in xenograft tissues was determined by qRT-PCR. (F) CDK6 expression in xenograft tissues was determined by Western blot. (G) Schematic diagram of LINC00473/miR-637/CDK6 axis in glioma. *p < .05.
Discussion
Glioma is challenging for the neurosurgeon due to its rapid proliferation and infiltrative growth, leading to invasion into normal brain tissue, and resulting in incomplete resection and recurrence [Citation18]. Recently, increasing evidence showed that lncRNAs contributed to glioma tumorigenesis and progression. For example, Gong et al. found that lncRNA CASC7 suppressed glioma proliferation and invasion through regulating Wnt/β-catenin pathway [Citation19]. Xu et al. suggested that lncRNA MEG3 reduced glioma cells proliferation and migration via regulation of Sirt7 and PI3K/AKT/mTOR pathway [Citation20]. Shen et al. found that lncRNA FOXD2-AS1 promoted glioma tumorigenesis via targeting the miR-185-5p/CCND2 axis [Citation21].
In the present study, we determined the roles of LINC00473 in glioma. QRT-PCR showed that LINC00473 expression was upregulated in glioma tissues and cell lines. High LINC00473 expression was correlated with advanced WHO grade, and low KPS score. Patients displaying a high expression of LINC00473 contributed to poor OS and DFS. LINC00473 inhibition suppressed glioma cells proliferation, invasion and EMT processes in vitro and reduced tumor growth in vivo. These data demonstrated that LINC00473 could act as a oncogenic lncRNA in glioma tumorigenesis.
Increasing evidence showed that cytoplasmic lncRNAs mainly function as ceRNAs or molecular sponges in carcinogenesis, including glioma [Citation22]. For example, Liu et al. showed that lncRNA HOTAIR promoted glioma progression by acting as a molecular sponge of miR-126-5p [Citation23]. Chen et al. found that HOXD-AS1 promoted metastasis in glioma via miR-130/E2F8 axis [Citation24]. Therefore, we speculated that LINC00473 might act as a ceRNA in glioma progression. In the current study, we showed that the majority of LINC00473 is located in cytoplasm, and bioinformatics analysis indicated that miR-637 could bind to LINC00473. The correlation between LINC00473 and miR-637 was further confirmed by Dual-luciferase reporter and RIP assays. Rescue assays showed that the effects of LINC00473 inhibition on glioma cells progressionn could be abolished by miR-637 inhibitors. Moreover, miR-637 expression was negatively associated with LINC00473 expression in glioma tissues. Thus, we suggested that LINC00473 exerted its oncogenic roles in glioma progression at least partly by regulating miR-637 expression.
CDK6 is a kinase catalytic subunit of a protein kinase complex that participates in G1 progression and G1/S transition [Citation25]. Recent studies showed that CDK6 could play critical roles in tumor progression. For example, Deng et al. found that miR-218 suppressed gastric cancer cell cycle progression through the CDK6/Cyclin D1/E2F1 axis [Citation26]. Lu et al. found that lncRNA NNT-AS1 promoted metastasis in hepatocellular carcinoma by the miR-363/CDK6 axis [Citation27]. In the present study, we showed that CDK6 expression was significantly increased in glioma and associated with poor OS and DFS. Subsequently, CDK6 was confirmed to be a target gene of miR-637 in glioma cells. Furthermore, we showed that LINC00473 suppression reduced CDK6 expression, and the effects could be partially abolished by miR-637 inhibitors. In addition, LINC00473 expression was positively correlated with CDK6 expression in glioma tissues. Thus, we suggested that LINC00473 promoted glioma proliferation and invasion by sponging miR-637 and upregulating CDK6.
Conclusions
Our study revealed that high LINC00473 expression promoted the proliferation and invasion of glioma cells by regulating miR-637/CDK6 axis, which provided a potential therapeutic target for glioma treatment
Declarations
Ethics approval and consent to participate
Ethical approval was given by the Ethics Committee of The First Affiliated Hospital of Zhengzhou University.
Disclosure statement
No potential conflict of interest was reported by the authors.
Availability of data and materials
The dataset supporting the conclusions of this article is included within the article.
References
- Schwartzbaum JA, Fisher JL, Aldape KD, et al. Epidemiology and molecular pathology of glioma. Nat Rev Neurol. 2006;2(9):494.
- McNeill KA. Epidemiology of brain tumors. Neurol Clin. 2016;34(4):981–998.
- Morgan LL. The epidemiology of glioma in adults: a “state of the science” review. Neuro-oncology, 2015;17(4):623–624.
- Feng X, Yu Y, He S, et al. Dying glioma cells establish a proangiogenic microenvironment through a caspase 3 dependent mechanism. Cancer Lett. 2017;385:12–20.
- Hanif F, Muzaffar K, Perveen K, et al. Glioblastoma multiforme: a review of its epidemiology and pathogenesis through clinical presentation and treatment. Asian Pac J Cancer Prev. 2017;18(1):3.
- Feinberg AP, Koldobskiy MA, Göndör A. Epigenetic modulators, modifiers and mediators in cancer aetiology and progression. Nat Rev Genet. 2016;17(5):284.
- Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10(3):155.
- Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10(1):38.
- Calle AS, Kawamura Y, Yamamoto Y, et al. Emerging roles of long non‐coding RNA in cancer. Cancer Sci. 2018;109(7):2093.
- Zhang H, Yang F, Chen S J, et al. Upregulation of long non-coding RNA MALAT1 correlates with tumor progression and poor prognosis in clear cell renal cell carcinoma. Tumor Biol. 2015;36(4):2947–2955.
- Liu H, Zhen Q, Fan Y. LncRNA GHET1 promotes esophageal squamous cell carcinoma cells proliferation and invasion via induction of EMT. Int J Biol Markers. 2017;32(4):403–408.
- Zhang G, An X, Zhao H, et al. Long non-coding RNA HNF1A-AS1 promotes cell proliferation and invasion via regulating miR-17-5p in non-small cell lung cancer. Biomed Pharmacother. 2018;98:594–599.
- Chen Z, Li JL, Lin S, et al. cAMP/CREB-regulated LINC00473 marks LKB1-inactivated lung cancer and mediates tumor growth.J Clin Invest. 2016;126(6):2267–2279.
- Zhu S, Fu W, Zhang L, et al. LINC 00473 antagonizes the tumour suppressor miR‐195 to mediate the pathogenesis of Wilms tumour via IKKα. Cell Prolif. 2018;51(1):e12416.
- Shi C, Yang Y, Yu J, et al. The long noncoding RNA LINC00473, a target of microRNA 34a, promotes tumorigenesis by inhibiting ILF2 degradation in cervical cancer. Am J Cancer Res. 2017;7(11):2157.
- Thomson D W, Dinger M E. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet. 2016;17(5):272.
- Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350.
- Chaichana KL, McGirt MJ, Laterra J, et al. Recurrence and malignant degeneration after resection of adult hemispheric low-grade gliomas. J Neurosurg. 2010;112(1):10–17.
- Gong X, Liao X, Huang M. LncRNA CASC7 inhibits the progression of glioma via regulating Wnt/β-catenin signaling pathway. Pathol-Res Pract. 2019;215:564–570.
- Xu D H, Chi G N, Zhao C H, et al. Long noncoding RNA MEG3 inhibits proliferation and migration but induces autophagy by regulation of Sirt7 and PI3K/AKT/mTOR pathway in glioma cells. J Cell Biochem. 2018. [Epub ahead of print].
- Shen F, Chang H, Gao G, et al. Long noncoding RNA FOXD2‐AS1 promotes glioma malignancy and tumorigenesis via targeting miR‐185‐5p/CCND2 axis. J Cell Biochem. 2019;120:9324–9336.
- Qin N, Tong GF, Sun LW, et al. Long noncoding RNA MEG3 suppresses glioma cell proliferation, migration, and invasion by acting as a competing endogenous RNA of miR-19a. Oncol Res. 2017;25(9):1471–1478.
- Liu L, Cui S, Wan T, et al. Long non‐coding RNA HOTAIR acts as a competing endogenous RNA to promote glioma progression by sponging miR‐126‐5p. J Cell Physiol. 2018;233(9):6822–6831.
- Chen Y, Zhao F, Cui D, et al. HOXD‐AS1/miR‐130a sponge regulates glioma development by targeting E2F8. Int J Cancer. 2018;142(11):2313–2322.
- Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 1999;13(12):1501–1512.
- Deng M, Zeng C, Lu X, et al. miR-218 suppresses gastric cancer cell cycle progression through the CDK6/Cyclin D1/E2F1 axis in a feedback loop. Cancer Lett. 2017;403:175–185.
- Lu YB, Jiang Q, Yang MY, et al. Long noncoding RNA NNT-AS1 promotes hepatocellular carcinoma progression and metastasis through miR-363/CDK6 axis. Oncotarget. 2017;8(51):88804.
