Abstract
Pancreatic cancer (PC) is a highly lethal human cancer. We previously found that Serine protease 3 (PRSS3), as an oncogene, is significantly upregulated in PC. In this study, we aimed to investigate the potential mechanism of PRSS3 dysregulation in PC. In this research, low miR-217 and high circ-ADAM9 expression were found in PC tissues and cell lines, which was closely associated with advanced clinical stage and lymph node metastasis. Patients with low miR-217 or high circ-ADAM9 expression had shorter survival time than those with high miR-217 or low circ-ADAM9 expression. Functionally, manipulation of miR-217 and circ-ADAM9 expression showed opposite effects on cell proliferation, migration and invasion. Stepwise mechanism studies indicated that circ-ADAM9 alleviated the inhibitory effect of miR-217 on PRSS3 by directly sponging miR-217 to increase the expression level of PRSS3, resulting in the activation of ERK/VEGF signalling pathway. In vivo, circ-ADAM9 silencing or miR-217 overexpression evidently retarded the growth of tumour, and the combination of them exhibited an additive inhibitory effect on tumourigenicity. Briefly, the ceRNA regulatory network of circ-ADAM9/miR-217/PRSS3 plays a pivotal role in PC progression by the regulation of ERK/VEGF signalling pathway.
Introduction
Pancreatic cancer (PC), a relatively rare malignant tumour of the digestive tract, is the fourth leading cause of cancer-associated deaths worldwide with increasing morbidity and mortality in recent years [Citation1]. Currently, surgical resection is still considered as the only potential effective treatment for PC patients, despite 50–80% of patients relapse within 1 year after surgery [Citation2]. The 5-year survival rate of patients with PC is extremely low (less than 5%) owing to most patients have no symptoms until the disease progresses to advanced stage, which makes them unsuitable for surgery and can only receive adjuvant systemic chemotherapy and radiotherapy [Citation3,Citation4]. As a result, a better understanding of the key factors that facilitate the development of PC will improve the early diagnosis and treatment, thereby prolonging the survival time of patients with PC.
Serine protease 3 (PRSS3), also known as mesotrypsinogen, is one of the major subtypes of trypsinogen that also contains PRSS1 and PRSS2. It is mainly synthesized in pancreatic acinar cells, which promotes digestion by secretion into the small intestine [Citation5]. Recent studies suggest that PRSS3 is aberrantly expression in human cancers and participates in the development and progression of cancer by acting as an oncogene or a tumour suppressor. For example, PRSS3 was identified to be downregulated in non-small cell lung cancer and hepatocellular carcinoma [Citation6,Citation7]. On the contrary, high PRSS3 was found in epithelial ovarian cancer and breast cancer [Citation8,Citation9], implying that PRSS3 functions in a cell- or disease-context-dependent manner. Likewise, we previously observed that PRSS3 was remarkably increased in PC and promoted PC growth and metastasis by activating ERK/VEGF pathway [Citation10]. However, the underlying mechanism of dysregulation of PRSS3 in PC remains unknown.
Circular RNA (circRNA), a special type of non-coding RNA, possesses a covalent closed loop structure without 5′-end cap and 3′-end ployA tail [Citation11]. The first circRNA was discovered in the early 1990s and has since been considered as a by-product of transcription [Citation12]. With the advent of high-throughput sequencing, a large number of circRNAs have been identified in human genome with highly stability and conservation [Citation13,Citation14]. Emerging evidence shows that circRNA can serve as a competing endogenous RNA (ceRNA) to regulate gene expression by sponging microRNAs (miRNAs) [Citation15]. For instance, circ-HIPK3 abundantly sponged miR-7 to increase the expression of miR-7 target genes, leading to promoting malignant progression of colorectal cancer [Citation16]. Hu et al. reported that circ-PRKCI enhanced cervical cancer growth and metastasis by sponging miR-545 to elevate EIF3C [Citation17]. Circ-MTO1 upregulates p21 expression by acting as a sponge of oncogenic miR-9 to inhibit hepatocellular carcinoma progression [Citation18]. However, little is known about the role of ceRNA regulatory network of circ-RNA/miRNA/gene in PC carcinogenesis.
In this study, we explored the underlying mechanism of PRSS3 dysregulation, and found that PRSS3 was controlled by circ-ADAM9 and miR-217 in PC. We also characterized the clinical and functional significance of circ-ADAM9 and miR-217 and elucidated the important role of circ-ADAM9/miR-217/PRSS3 ceRNA regulatory network in the progression of PC.
Materials and methods
Patients and cell lines
This study was carried out with the approval of the ethics committee of the first affiliated hospital of Zhengzhou University. A total of 58 pairs of fresh frozen PC and matched normal tissues were collected from the first affiliated hospital of Zhengzhou University. Patients who received preoperative anti-neoplastic treatment were excluded in our study. All patients provided informed consent before starting this study. Patients were followed up regularly by telephone or e-mail after surgery. All human PC cell lines including PANC1, MiaPaca2, SW1990, Capan1 and BxPC3 and a normal pancreatic ductal epithelial cell line HPDE were routinely cultured in RPMI1640 or DMEM complete medium plus 10% foetal bovine serum (FBS) and 1% penicillin and streptomycin in the incubator at 37 °C with 5% CO2.
Oligonucleotides, plasmids and transfection
All oligonucleotides including miR-217 mimics or inhibitors, small interfering RNAs targeting circ-ADAM9 (si-circ-ADAM9) and PRSS3 (si-PRSS3) and their corresponding controls were commercially obtained from Gene-Pharma Company (Shanghai, China). The expression vectors of circ-ADAM9 (pCD-ciR) and PRSS3 (pcDNA3.0) were purchased from Geneseed Company (Guangzhou, China) and Invitrogen Corporation (Carlsbad, MA), respectively. Besides, the cholesterol-conjugated miR-217 mimics and si-circ-ADAM9 (Gene-Pharma) were employed for the in vivo injection. All above constructs were alone or in combination transfected into MiaPaca2 and Capan1 cells using Opti-MEM and Lipofectamine 2000 based on Manufacturer's manual (Invitrogen). After transfection for 48 h, cells in each well were washed and collected. The transfection efficiency was detected by quantitative reverse transcription PCR (qRT-PCR) or western blot.
Dual luciferase reporter assay
The full-length sequences of PRSS3 3′-UTR and circ-ADAM9 with or without mutated predicted miR-217 binding site were subcloned into pmirGLO reporter vector (Promega, Madison, WI) between SpeI and Hind III restriction sites, respectively. Then, the above luciferase reporter vectors were cotransfected with miR-217 or control mimics into MiaPaca2 and Capan1 cells with Lipofectamine 2000 (Invitrogen). Forty-eight hours later, cells in each well were collected and the luciferase activity was measured by a luciferase assay kit (BioVision. Inc., Milpitas, CA). Renilla luciferase was used as a control luciferase reporter for normalization and selection.
Quantitative reverse transcription PCR (qRT-PCR)
Total RNA was extracted from PC cell lines and tissues using Trizol solution (Invitrogen, Carlsbad, MA) and dissolved with 30 μl of enzyme-free water. Next, 1 μg extracted RNA was utilized for reverse transcription to obtain the single-stranded cDNA. Finally, quantitative PCR analyses were performed in triplicate with 35 cycles using a SYBR Green Mix kit (Takara, Otsu, Japan). U6 and GAPDH were used as internal references for miRNAs and circRNAs/mRNAs, respectively. The sequences of primers used in this study are listed below:
Western blot
The protocol of western blot was described in our previous study [Citation10]. The primary antibodies used in this study were anti-PRSS3 (#ab107430, Abcam, Cambridge, UK), anti-p-ERK (#4370, Cell Signaling Technology, Danvers, MA), anti-ERK (#4695, Cell Signaling Technology, MA, USA), anti-VEGF (#sc-152, Santa Cruz Biotechnology, Santa Cruz, TX), and anti-GAPDH (#ab181602, Abcam, Cambridge, UK) antibodies. GAPDH was used as internal reference. The grey values of western blot results were analyzed by Image J software.
Cell function assays
The proliferation and migration/invasion of PC cells were assessed by cell counting cit-8 (CCK-8) and transwell assays, respectively. For CCK-8 assay, MiaPaca2 and Capan1 cells were respectively plated into 96-well plates and incubated for 0 h, 24 h, 48 h, 72 h and 96 h. Subsequently, 10 μl of CCK-8 reagent (Dojindo Laboratories, Kumamoto, Japan) was added into each well and incubated for another 2 h at 37 °C, followed by a record of the absorbance at 450 nm. For transwell assay, 100 μl of MiaPaca2 and Capan1 cells were, respectively, seeded into transwell chambers (Corning, NY) coated with (invasion) or without (migration) the matrigel. Sixteen hours later, the migrated and invaded cells on the surfaces of lower chambers were fixed with methanol and stained with crystal violet, followed by analysis of the ability of cells to invade and migrate.
RNA pull-down assay
The biotin-labelled miR-217 and control mimics were commercially purchased from Gene-Pharma Company (Shanghai, China) and then transfected into MiaPaca2 and Capan1 cells using Lipofectamine 2000 (Invitrogen, Carlsbad, MA). 48 h later, cells were collected and incubated with streptavidin magnetic beads (Invitrogen, Carlsbad, MA) for 2 h at room temperature on the rotary shaker. Besides, 50 μl of cell lysate was aliquot for input. Then, the miR-217-binding RNAs were isolated using Trizol solution (Invitrogen, Carlsbad, MA), followed by qRT-PCR analysis for the expression levels of circ-ADAM9, circ-ATAD1, circ-SYPL1, circ-SNF8 and circ-ZCCHC14.
In vivo xenograft model
The procedures of animal experiment in our study were in agreement with the Animal Policy and Welfare Committee of the first affiliated hospital of Zhengzhou University. All 4–6-week-old male BALB/c nude mice were obtained from Chinese Academy of Science (Shanghai, China) and routinely grown under specific pathogen-free condition. 3 × 106 Capan1 cells were subcutaneously injected into nude mice to establish the xenograft model. One week after the injection, the volume of the subcutaneous tumours reached approximately 100 cm3. Then, the nude mice were randomly divided into four groups (si-NC, si-circ-ADAM9, miR-217 and si-circ-ADAM9 + miR-217), and the corresponding cholesterol-conjugated oligonucleotides were intratumourally injected into nude mice twice or third weekly (n = 5 for each group). Five weeks later, all nude mice were sacrificed by cervical dislocation and the tumour tissues in each group were collected and weighed.
Statistical analyses
All biological experiments in this study were effectively repeated at least three times. Data were represented as mean ± standard deviation (SD). Student’s t or Chi-square test was employed to determine the differences between two groups. The correlation was tested by Pearson’s correlation coefficient. And, the survival curves of PC patients with low/high circ-ADAM9/miR-217 expression were calculated using the Kaplan–Meier plot and the long-rank test. All statistical results were two-sided and analyzed with SPSS 22.0 (SPSS Inc., Chicago, IL) or GraphPad 7.0 software (GraphPad Software, La Jolla, CA). *p < .05, **p < .01 and ***p < .001.
Results
miR-217 is the PRSS3-related miRNA in PC
In our previous study, we found that PRSS3 was markedly increased in PC cell lines and tissues and linked to aggressive characteristics [Citation10]. To explore the underlying mechanism of PRSS3 dysregulation in PC, we first analyzed the TCGA database. The results showed that gene mutation (less than 1%) (), copy number variation () and epigenetic modification () were not responsible for PRSS3 overexpression in PC. We then focused on the post-transcriptional regulation of PRSS3. By searching for the Targetscan and miRanda online database, the complementary pairing between the sequences of PRSS3 3′-UTR and miR-217 was observed (. The luciferase reporter assay results showed that overexpression of miR-217 significantly decreased the luciferase activity of wild-type vector, but had no effect on the mutant one (), suggesting that the “UGCAGU” motif on PRSS3 3′-UTR is the site of action of miR-217 targeting PRSS3. Next, we investigated the expression level of miR-217 in PC and its clinical significance. We found that miR-217 was dramatically downregulated in PC tissues as compared with adjacent normal tissues (. And, low miR-217 was frequently observed in PC patients with advanced clinical stage () and lymph node metastasis (. More strikingly, PC patients with low miR-217 displayed shorter survival time than those with high miR-217 (. These above results imply that miR-217 is an important dysregulated miRNA in PC and its downregulation may be responsible for PRSS3 upregulation.
Figure 1. miR-217 targets the 3′-UTR of PRSS3 in PC cells. (A) The mutant status of PRSS3 in PC patients from TCGA database. (B) The mRNA expression of PRSS3 in PC patients with different types of copy number variations. (C) The correlation between PRSS3 expression and methylation level in PC tissues. (D) The diagrammatic sketch showing the binding site between 3′-UTR of PRSS3 and miR-217. (E) The relative luciferase activities of wild-type and mutated PRSS3 3′-UTR luciferase vectors co-transfected with control or miR-217 mimics in MiaPaca2 cells. (F) qRT-PCR analysis for miR-217 expression in PC and matched normal tissues. (G, H) qRT-PCR analysis for miR-217 expression in PC patients with different TNM stage (I and II versus III and IV) (G) and lymph node status (no metastasis versus metastasis) (H). (I) The overall survival curves of PC patients with low and high miR-217 expression. UTR: untranslated region; LN: lymph node; ***p < .001.
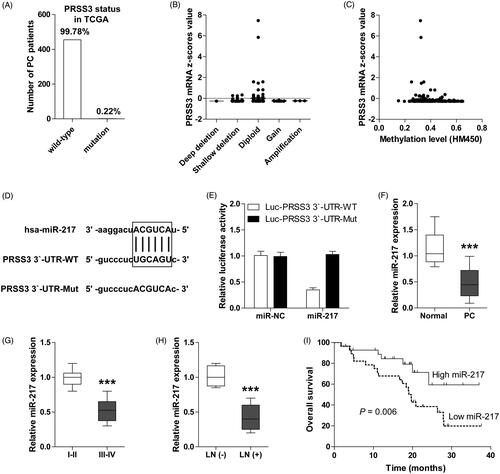
miR-217 inhibits the proliferation, migration and invasion of PC cells by targeting PRSS3
Consistently, miR-217 expression was evidently decreased in PC cell lines when compared with normal pancreatic ductal epithelial HPDE cell (. Due to MiaPaca2 and Capan1 exhibited relatively high and low expression of miR-217, respectively, we chose MiaPaca2 for miR-217 silencing and Capan1 for miR-217 overexpression. The transfection efficiencies were confirmed by qRT-PCR (). We found that knockdown of miR-217 increased, while overexpression of miR-217 decreased both the mRNA and protein expression of PRSS3 in PC cells (). Moreover, miR-217 expression was strongly inversely correlated with PRSS3 expression in PC tissues (. Functionally, miR-217-silencing MiaPaca2 cells showed strong proliferative, migrative and invasive capacities compared to control cells; however, the aggressive phenotypes promoted by miR-217 knockdown could be evidently blocked by PRSS3 knockdown (). On the contrary, ectopic expression of miR-217 significantly attenuated the ability of Capan1 cells to proliferate, migrate and invade, and these inhibitory effects could be effectively rescued by PRSS3 overexpression (). In all, these data indicate that miR-217 exerts the tumour-suppressive effect mainly by targeting oncogenic PRSS3 in PC.
Figure 2. miR-217 inhibits the aggressive phenotype of PC cells by targeting PRSS3. (A) qRT-PCR analysis for miR-217 expression in PC cell lines. (B, C) qRT-PCR analysis for miR-217 and PRSS3 expression in miR-217-knockdown MiaPaca2 (B) and miR-217-overexpressing Capan1 cells (C). (D) Western blot analysis for PRSS3 expression in miR-217-knockdown/overexpressing MiaPaca2/Capan1 cells. GAPDH was used as a control reference. (E) The correlation between miR-217 and PRSS3 expression in PC tissues. (F, G) CCK-8 proliferation assays in miR-217-knockdown MiaPaca2 cells (F) or in miR-217-overexpressing Capan1 cells (G) transfected with PRSS3 small interfering RNA or PRSS3 expression plasmid. (H,I) Transwell migration and invasion assays in miR-217-knockdown MiaPaca2 cells or in miR-217-overexpressing Capan1 cells transfected with PRSS3 small interfering RNA (H) or PRSS3 expression plasmid (I). **p < .01.
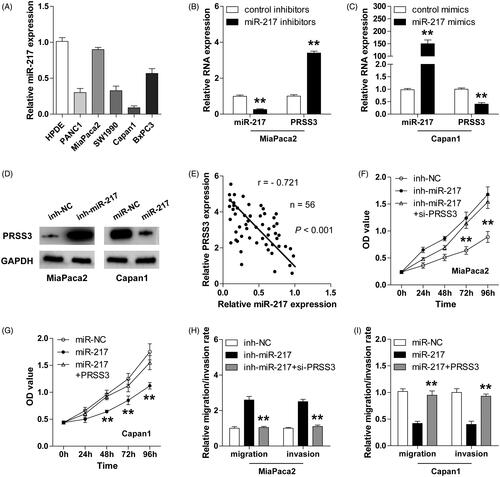
Circ-ADAM9 serves as a sponge of miR-217 in PC
Accumulating evidence suggests that circRNA is capable to sponge miRNAs to regulate gene expression [Citation19]. To identify the miR-217-associated circRNAs, we analyzed the starBase V2.0 online database and found several circRNAs that were predicted to interact with miR-217. The top five circRNAs were shown in according to the clipReadNum values. Further, the results of RNA pull-down assays showed that circ-ADAM9, but not circ-ATAD1, circ-SYPL1, circ-SNF8, circ-ZCCHC14, was abundantly enriched by miR-217 probe both in MiaPaca2 and Capan1 cells (). Then, we constructed the luciferase vector of circ-ADAM9 with wide-type or mutated miR-217 binding site to conduct luciferase reporter assay (), the results showed that miR-217 overexpression significantly reduced the luciferase activity of wide-type vector, nevertheless, the inhibitory action was almost completely abrogated after the mutation of miR-217 binding site (). Moreover, overexpression and ablation of circ-ADAM9 evidently decreased and increased the expression level of miR-217, respectively (). And, a strong negative correlation between circ-ADAM9 and miR-217 was identified in PC tissues (. Taken together, these results demonstrate that miR-217 is sponged and inhibited by circ-ADAM9 in PC.
Figure 3. miR-217 is absorbed and inhibited by circ-ADAM9 in PC cells. (A) The top five miR-217-associated circRNAs predicted by starBase V2.0 online program. (B,C) RNA pull-down assay in MiaPaca2 (B) and Capan1 (C) cells transfected with biotin-labelled miR-217 or control probe, followed by qRT-PCR analysis for the expression of the indicated circRNAs. (D) The diagrammatic sketch showing the binding site between circ-ADAM9 and miR-217. (E,F) The relative luciferase activities of wild-type and mutated circ-ADAM9 luciferase vectors co-transfected with control or miR-217 mimics in MiaPaca2 (E) and Capan1 (F) cells. (G,H) qRT-PCR analysis for the expression of circ-ADAM9 and miR-217 in circ-ADAM9-overexpressing MiaPaca2 (G) and circ-ADAM9-silencing Capan1 cells (H). (I) The correlation between circ-ADAM9 and miR-217 expression in PC tissues. **p < .01.
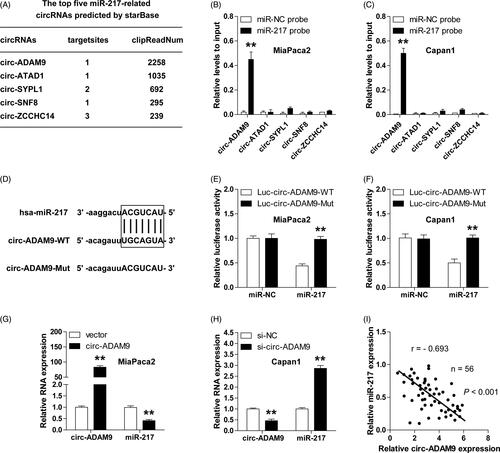
Circ-ADAM9 is remarkably upregulated in PC and predicts poor outcome
We next assessed the expression level and clinical significance of circ-ADAM9 in PC. As shown in , circ-ADAM9 expression was uniformly increased in PC cell lines. Similarly, high circ-ADAM9 was observed in PC tissues compared with matched non-cancerous tissues (), which was closely related to advanced TNM stage and lymph node metastasis (). More importantly, the survival time of PC patients with high circ-ADAM9 expression was significantly shorter than that of patients with low circ-ADAM9 expression (. These findings reveal that circ-ADAM9 is also a key dysregulated gene in PC.
Figure 4. Overexpressed circ-ADAM9 is identified in PC cell lines and tissues and predicts poor outcome. (A,B) qRT-PCR analysis for the expression of circ-ADAM9 in PC cell lines (A) and tissues (B). (C,D) qRT-PCR analysis for circ-ADAM9 expression in PC patients with different TNM stage (I and II versus III and IV) (C) and lymph node status (no metastasis versus metastasis) (D). (E) The overall survival curves of PC patients with low and high circ-ADAM9 expression. LN: lymph node; ***p < .001.
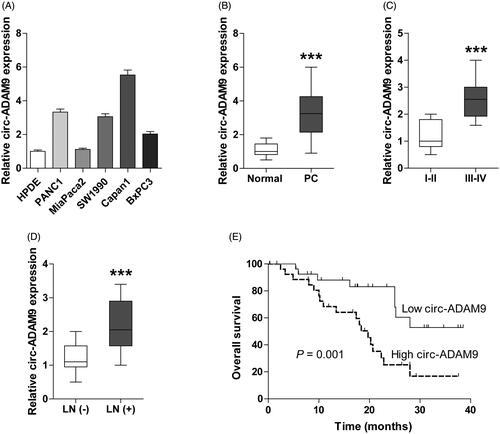
Circ-ADAM9/miR-217/PRSS3 axis promotes PC progression by regulating ERK/VEGF signalling
Subsequently, we wondered whether circ-ADAM9 affected the expression of miR-217 target gene PRSS3 by adsorption of miR-217. The qRT-PCR and western blot results showed that enforced expression of circ-ADAM9 notably increased the mRNA and protein expression of PRSS3, however, this increase was offset by miR-217 overexpression (). Contrarily, circ-ADAM9 knockdown dramatically decreased PRSS3 expression, and this inhibition could be distinctly rescued by miR-217 depletion (). We previously found that PRSS3 was capable to activate ERK/VEGF pathway [Citation10]. Consistently, circ-ADAM9 overexpression and knockdown significantly increased and decreased phosphorylated ERK (pERK) and VEGF expression in PC cells, respectively, and these effects were also abolished by altering the expression of miR-217 (). Moreover, exogenous expression of circ-ADAM9 markedly promoted the proliferation as well as migration and invasion of MiaPaca2 cells, and miR-217 overexpression or PRSS3 knockdown evidently blocked the above facilitating effects (). In contrast, the malignant phenotypic inhibition caused by circ-ADAM9 silencing could be effectively rescued by miR-217 ablation or PRSS3 overexpression ().
Figure 5. The ceRNA network of circ-ADAM9/miR-217/PRSS3 axis contributes to the progression of PC through activating ERK/VEGF signalling pathway. (A,B) qRT-PCR analysis for PRSS3 mRNA expression in circ-ADAM9-overexpressing MiaPaca2 or circ-ADAM9-silencing Capan1 cells co-transfected with miR-217 mimics (A) or inhibitors (B). (C,D) Western blot analysis for the protein expression of PRSS3, pERK, ERK, VEGF in circ-ADAM9-overexpressing MiaPaca2 or circ-ADAM9-silencing Capan1 cells transfected with miR-217 mimics (C) or inhibitors (D). (E,G) CCK-8 proliferation (E) and transwell (G) assays in circ-ADAM9-overexpressing MiaPaca2 cells with or without miR-217 overexpression or PRSS3 knockdown. (F,H) CCK-8 proliferation (F) and transwell (H) assays in circ-ADAM9-silencing Capan1 cells with or without PRSS3 overexpression or miR-217 knockdown. (I,J) The volume (I) and weight (J) of tumours in the indicated groups. The black arrows denote the time and number of specific intratumoural injections of cholesterol-conjugated oligonucleotides. *p < .05, **p < .01 and ***p < .001.
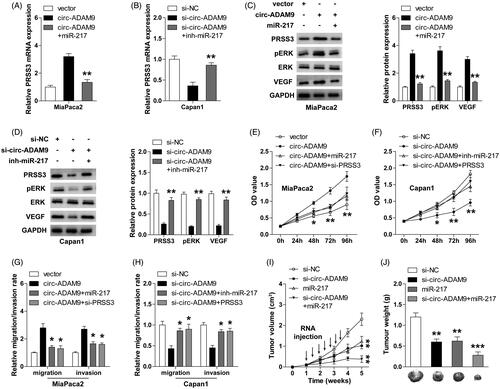
Circ-ADAM9 silencing or miR-217 overexpression effectively delayed tumour growth in vivo
To test whether circ-ADAM9 and miR-217 also functioned in vivo, we established a xenograft model by subcutaneous injection of Capan1 cells into nude mice. The cholesterol-conjugated oligonucleotides (si-NC, si-circ-ADAM9, miR-217 and si-circ-ADAM9 + miR-217) were intratumourally injected after the subcutaneous tumours reached approximately 100 cm3 (. Thirty-five days later, the results showed that the volume and weight of tumours in si-circ-ADAM9 or miR-217 group were significantly smaller than those in control group (). Moreover, the combination of si-circ-ADAM9 and miR-217 displayed an additive inhibitory effect on tumour growth (). These data indicate that circ-ADAM9 and miR-217 are an oncogene and a tumour suppressor in vivo, respectively, which is consistent with clinical and in vitro findings.
Discussion
We previously identified that PRSS3 was frequently increased in PC and its upregulation implicated a link with aggressive properties [Citation10]. In this study, we explored the underlying mechanism of PRSS3 dysregulation. We found that PRSS3 upregulation was attributed to miR-217 decrease and circ-ADAM9 increase in PC. Overexpression of miR-217 and circ-ADAM9 respectively inhibited and promoted the proliferative, migrative and invasive capacities of PC cells, however, knockdown of miR-217 or circ-ADAM9 showed the opposite trend. Subsequent mechanism investigations revealed that miR-217 could target the 3′-UTR of PRSS3 mRNA to inhibit PRSS3 expression, however, circ-ADAM9 was capable to act as a sponge of miR-217 to alleviate the inhibitory effect of miR-217 on PRSS3, resulting in the activation of ERK/VEGF signalling, thus promoting the aggressive progression of PC. Therefore, our data highlight the importance of circ-ADAM9/miR-217 axis in regulating PRSS3 and also provide novel insights into PC tumourigenesis.
PRSS3 is an atypical subtype of trypsinogen that has been implicated to be aberrantly expressed in different tumours, including PC [Citation10,Citation20,Citation21]. However, little is known about the underlying mechanisms of PRSS3 dysregulation. Marsit et al. reported that PRSS3 was dramatically decreased in non-small cell lung cancer due to promoter hypermethylation [Citation7], suggesting that epigenetic silencing mechanisms may be responsible for the dysregulation of PRSS3. However, neither epigenetic nor genetic alteration was evidently observed in PRSS3 of PC tissues from TCGA database, implying that PRSS3 functions in a cell- or disease-context-dependent manner and post-transcriptional regulation may be involved in PRSS3 aberration. It has been extensively reported that miRNA can tightly regulate the expression levels of its downstream target genes through promoting the degradation or inhibiting translation by completely or incompletely pairing with the 3′-UTR sequence of target genes [Citation22]. Herein, we also identified a miRNA, miR-217, which bound to 3′-UTR of PRSS3 to decrease the expression of PRSS3. Many studies have reported that miR-217 was downregulated in human cancers, including gastric cancer [Citation23], osteosarcoma [Citation24], hepatocellular carcinoma [Citation25], renal cell carcinoma [Citation26, Citation27], PC [Citation28], acute myeloid leukaemia [Citation29] and triple-negative breast cancer [Citation30]. Consistent with the above studies, we found that miR-217 was lowly expressed in PC, which closely associated with dismal prognosis. Moreover, miR-217-induced tumour-inhibiting effect could be mostly blocked by PRSS3. Altogether, these indicate that miR-217 is an upstream negative regulator of PRSS3 and acts as a tumour suppressor by targeting PRSS3 in PC.
Multiple lines of evidence unveil that circRNA, a special type of non-coding RNA, can function as a ceRNA to regulate gene expression by sponging miRNAs, thus, participating in the occurrence and progression of cancer [Citation31,Citation32]. In the present study, we found that miR-217 was abundantly absorbed by circ-ADAM9, as determined by RNA pull-down and luciferase reporter assays. Overexpression and knockdown of circ-ADAM9 significantly decreased and increased miR-217 expression in PC cells, respectively, indicating that circ-ADAM9 is an upstream negative regulator of miR-217. Furthermore, circ-ADAM9 could remarkably elevated PRSS3 expression and subsequently activated the ERK/VEGF pathway to facilitate the growth and metastasis of PC, but this oncogenic effect was partially abolished by miR-217, suggesting that miR-217 is an important mediator of circ-ADAM9 and PRSS3. Further research is warranted to investigate whether this circ-ADAM9/miR-217/PRSS3 ceRNA axis also functions in other malignancies. Of note, silencing of circ-ADAM9 or ectopic expression of miR-217 markedly retarded the in vivo tumour growth, and the combination of them had an additive inhibitory effect, implying that depletion of circ-ADAM9 and restoration of miR-217 may be a promising treatment for PC.
Conclusion
PRSS3 is regulated by circ-ADAM9/miR-217 axis, and the ceRNA regulatory network of circ-ADAM9/miR-217/PRSS3 plays an essential role in the development and progression of PC through the regulation of ERK/VEGF signalling. Targeting of this axis may be an efficacious approach against this lethal disease.
| Abbreviations | ||
| ceRNA | = | competing endogenous RNA |
| circRNA | = | circular RNA |
| PC | = | pancreatic cancer |
| PRSS3 | = | Serine protease 3 |
| microRNA | = | miRNA |
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Kamisawa T, Wood LD, Itoi T, et al. Pancreatic cancer. Lancet. 2016;388(10039):73–85.
- Collisson EA, Bailey P, Chang DK, et al. Molecular subtypes of pancreatic cancer. Nat Rev Gastroenterol Hepatol. 2019;16(4):207.
- Strobel O, Neoptolemos J, Jager D, et al. Optimizing the outcomes of pancreatic cancer surgery. Nat Rev Clin Oncol. 2019;16(1):11–26.
- He C, Zhang Y, Cai Z, et al. Nomogram to predict cancer-specific survival in patients with pancreatic acinar cell carcinoma: a competing risk analysis. J Cancer. 2018;9(22):4117–4127.
- Tani T, Kawashima I, Mita K, et al. Nucleotide sequence of the human pancreatic trypsinogen III cDNA. Nucleic Acids Res. 1990;18(6):1631.
- Lin B, Zhou X, Lin S, et al. Epigenetic silencing of PRSS3 provides growth and metastasis advantage for human hepatocellular carcinoma. J Mol Med. 2017;95(11):1237–1249.
- Marsit CJ, Okpukpara C, Danaee H, et al. Epigenetic silencing of the PRSS3 putative tumor suppressor gene in non-small cell lung cancer. Mol Carcinog. 2005;44(2):146–150.
- Ma R, Ye X, Cheng H, et al. PRSS3 expression is associated with tumor progression and poor prognosis in epithelial ovarian cancer. Gynecol Oncol. 2015;137(3):546–552.
- Qian L, Gao X, Huang H, et al. PRSS3 is a prognostic marker in invasive ductal carcinoma of the breast. Oncotarget. 2017;8(13):21444–21453.
- Jiang G, Cao F, Ren G, et al. PRSS3 promotes tumour growth and metastasis of human pancreatic cancer. Gut. 2010;59(11):1535–1544.
- Li X, Yang L, Chen LL. The biogenesis, functions, and challenges of circular RNAs. Mol Cell. 2018;71(3):428–442.
- Nigro JM, Cho KR, Fearon ER, et al. Scrambled exons. Cell. 1991;64(3):607–613.
- Kristensen LS, Hansen TB, Veno MT, et al. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37(5):555–565.
- Qiu LP, Wu YH, Yu XF, et al. The emerging role of circular RNAs in hepatocellular carcinoma. J Cancer. 2018;9(9):1548–1559.
- Zhong Y, Du Y, Yang X, et al. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol Cancer. 2018;17(1):79.
- Zeng K, Chen X, Xu M, et al. CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell Death Dis. 2018;9(4):417.
- Hu C, Wang Y, Li A, et al. Overexpressed circ_0067934 acts as an oncogene to facilitate cervical cancer progression via the miR-545/EIF3C axis. J Cell Physiol. 2018;234(6):9225–9232.
- Han D, Li J, Wang H, et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66(4):1151–1164.
- Verduci L, Strano S, Yarden Y, et al. The circRNA-microRNA code: emerging implications for cancer diagnosis and treatment. Mol Oncol. 2019;13(4):669–680.
- Hockla A, Miller E, Salameh MA, et al. PRSS3/mesotrypsin is a therapeutic target for metastatic prostate cancer. Mol Cancer Res. 2012;10(12):1555–1566.
- Radisky ES. PRSS3/mesotrypsin in prostate cancer progression: implications for translational medicine. Asian J Androl. 2013;15(4):439–440.
- Saliminejad K, Khorram KH, Soleymani FS, et al. An overview of microRNAs: biology, functions, therapeutics, and analysis methods. J Cell Physiol. 2019;234(5):5451–5465.
- Dong X, He X, Guan A, et al. Long non-coding RNA Hotair promotes gastric cancer progression via miR-217-GPC5 axis. Life Sci. 2019;217:271–282.
- Wang B, Qu XL, Liu J, et al. HOTAIR promotes osteosarcoma development by sponging miR-217 and targeting ZEB1. J Cell Physiol. 2019;234(5):6173–6181.
- Wang H, Ke J, Guo Q, et al. Long non-coding RNA CRNDE promotes the proliferation, migration and invasion of hepatocellular carcinoma cells through miR-217/MAPK1 axis. J Cell Mol Med. 2018;22(12):5862–5876.
- Ding J, Yeh CR, Sun Y, et al. Estrogen receptor beta promotes renal cell carcinoma progression via regulating LncRNA HOTAIR-miR-138/200c/204/217 associated CeRNA network. Oncogene. 2018;37(37):5037–5053.
- Hong Q, Li O, Zheng W, et al. LncRNA HOTAIR regulates HIF-1alpha/AXL signaling through inhibition of miR-217 in renal cell carcinoma. Cell Death Dis. 2017;8(5):e2772.
- Karamitopoulou E, Haemmig S, Baumgartner U, et al. MicroRNA dysregulation in the tumor microenvironment influences the phenotype of pancreatic cancer. Mod Pathol. 2017;30(8):1116–1125.
- Yan J, Wu G, Chen J, et al. Downregulated miR-217 expression predicts a poor outcome in acute myeloid leukemia. CBM. 2018;22(1):73–78.
- Zhou W, Song F, Wu Q, et al. MiR-217 inhibits triple-negative breast cancer cell growth, migration, and invasion through targeting KLF5. PLoS One. 2017;12:e176395.
- Shang Q, Yang Z, Jia R, et al. The novel roles of circRNAs in human cancer. Mol Cancer. 2019;18(1):6.
- Abdollahzadeh R, Daraei A, Mansoori Y, et al. Competing endogenous RNA (ceRNA) cross talk and language in ceRNA regulatory networks: a new look at hallmarks of breast cancer. J Cell Physiol. 2018;234(7):10080–10100.
