Abstract
Background
Gastric cancer (GC) is a global leading source of cancer-associated deaths. Circular RNAs (circRNAs) are a new type of non-coding RNA and promising biomarkers for diagnosis of multiple diseases such as cancer.
Methods
Circ-PRMT5 expression was validated in 90 GC patient tissues and 6 different GC cells by qRT-PCR. Sublocalization of circ-PRMT5 in GC cells was determined in isolated nuclear and cytoplasmic RNAs. CircInteractome and miRanda were used to predict binding sites between circ-PRMT5 with micRNAs, and micRNAs with target mRNA. The correlation between genes was determined by the Pearson correlation analysis. The molecular mechanism was demonstrated by RNA in vivo precipitation, point mutation, luciferase activity and rescue experiments.
Results
Circ-PRMT5 expression was significantly higher in GC than in adjacent normal tissues, and GC patients with circ-PRMT5 high expression had shorter survival times. Functionally, circ-PRMT5 silence inhibited GC cell growth and invasion. Mechanism analysis showed that circ-PRMT5 sponged miR-145/miR-1304 to upregulate MYC expression and GC development.
Conclusion
Our findings demonstrated that circ-PRMT5 function as an oncogene in GC patients by targeting miR-145/miR-1304/MYC axis. High circ-PRMT5 expression may provide a poor prognostic indicator of survival in GC patients and targeting circ-PRMT5/miR-145/miR-1304/MYC axis may be a novel therapeutic strategy for GC.
Introduction
Gastric cancer (GC) remains to be a main hazard to human healthiness, the fourth most universal cancer and the third leading cause of global cancer-associated deaths as said by global cancer statistics [Citation1]. In spite of the application of various improvements in diagnosis and treatment, the GC prognosis continues to be quite poor, with a 5-year overall survival lower than 40% in many countries, due to the tumour invasion, metastasis and recurrence [Citation2,Citation3].
In the past decades, non-coding RNAs (ncRNAs), including microRNA (miRNA) and long non-coding RNA (lncRNA) have been found to be deregulated in GC patients, and have prospective clinical applications [Citation4,Citation5].
CircRNAs are a novel identified and distinct type of ncRNAs derived from introns, exons or intergenic regions, which is characterized by a covalently closed structure without 5′caps and 3′tails and protein coding ability. CircRNAs also exhibit tissue or cell-specific expression, and are conserved through species owing to their resistance to RNase R [Citation6–8]. Compared with linear RNAs, circRNAs are really stable and primarily in the cytoplasm [Citation9,Citation10]. CircRNAs comprise miRNA response elements (MREs) that can be applied to investigate miRNA-specific antagonists. More and more evidences indicate that circRNAs interact with RNA binding proteins (RBPs) and act as miRNA sponges to regulate gene expression [Citation11].
CircRNAs participate in many physiological and pathological procedures, such as transcription, RNA decay, mRNA splicing and translation, and their dysregulation results in abnormality of cellular functions and human illnesses [Citation12]. Especially, the tumourigenesis functions of circRNAs are widely explored recently. It is found that many circRNA types are aberrantly expressed in GC, HCC, oesophageal squamous cancer, CRC, oral cancer and bladder cancer and related with cancer progression [Citation13–17]. Therefore, it is crucial to recognize deregulated circRNAs, discern novel molecular mechanisms and therapeutic targets for GC treatment.
Recently, a novel circRNA, circ-PRMT5 (chr14:23395341–23396023), was reported to promote the metastasis of urothelial carcinoma of the bladder by sponging miR-30c to induce EMT [Citation18]. However, its role in GC is unknown.
In current study, we aimed to explore the function, molecular mechanisms and clinical significances of the circ-PRMT5 in GC. Our results validated that circ-PRMT5 expression level was dramatically increased in GC tissues. High expression of PRMT5 was associated with the poor prognosis of patients with GC. More importantly, we found that PRMT5 functioned as a sponge of miR-145 and miR-1304 to upregulate the MYC expression and consequently promoted the tumourigenesis of GC.
Methods
Clinical data and patient tissues
GC and paired adjacent normal gastric tissue samples from 90 GC patients were acquired from patients undergoing surgery at the Affiliated Hospital of Inner Mongolia University for Nationalities and Nantong Tumor Hospital. All specimens were snap frozen in liquid nitrogen upon collection and stored at −80 °C until use. All patients were regularly followed up and overall survival (OS) time was from the date of surgery to the date of death.
Consent to participate and ethics approval
All samples were collected with written informed consents from the patients before the study was initiated. All experiments were approved by the Ethics Committee of Second Affiliated Hospital of Kunming Medical University and informed in accordance with Declaration of Helsinki.
Reagents
Dulbecco’s modified Eagle’s medium (DMEM) was from Gibco (Rockford, MD). Fetal bovine serum (FBS) was from HyClone (Northbrook, IL). RNAiso Plus, PrimeScrip™ RT Master Mix (TaKaRa Dalian, China) and SYBR Green Premix Ex Taq™ II (Takara) were from TaKaRa (Dalian, China). MiRNeasy Mini Kit, miScript II RT Kit and miScript SYBR Green PCR Kit were from Qiagen (Duesseldorf, Germany). CCK-8 assay kit was from Dojindo Corp (Kyushu, Japan). Matrigel was from BD (Franklin Lakes, NJ). Dual-luciferase reporter assay system was from Promega (Beijing) Biotech Co., Ltd (Beijing, China). RNA subcellular isolation kit was from Active Motif China (Shanghai, China). RIPA lysis buffer was from Beyotime (Shanghai, China). SuperSignal West Dura Extended Duration Substrate was from Thermo Fisher (Chicago, IL).
Cell lines and cell culture
Six GC cell lines (AGS, MKN-28, MKN45, BGC823, MGC803, SGC7901) and a normal human gastric epithelial cell line (GES-1) were purchased from American Type Culture Collection (ATCC). The cells were cultured in DMEM containing 10% FBS, 100 μg/mL streptomycin, and 100 U/mL penicillin. All cells were cultured in a humidified atmosphere and 5% CO2 at 37 °C.
Analysis and target gene prediction
The sponged miRNAs by the circ-PRMT5 were predicted using the website tool CircInteractome (https://circinteractome.nia.nih.gov/index.html). The target mRNA of miR-145 and miR-1304 was predicted by the website tool miRanda (http://www.microrna.org/).
Quantitative real-time PCR (qRT-PCR)
Total RNA was isolated using RNAiso Plus. RNA purity and concentration were confirmed spectrophotometrically using the NanoDrop ND-1000 (Thermo Fisher Scientific, Wilmington, DE). OD260/OD230 ratios >1.8 but <2.1 were accepted. Reverse transcription was achieved using PrimeScrip™ RT Master Mix, and cDNA amplification was achieved using SYBR Green Premix Ex Taq™ II following the manufacturer’s instructions.
MiRNA was isolated using a miRNeasy Mini Kit, reverse transcription was achieved using a miScript II RT Kit, and cDNA amplification was achieved using a miScript SYBR Green PCR Kit following the manufacturer’s instructions. qRT-PCR was conducted on an AB7300 thermo recycler (Applied Biosystems, Carlsbad, CA) with primers and TaqMan Universal PCR Master Mix. GAPDH was used as the reference gene for mRNA and circRNA. U6 was used as an internal control for miRNA. Gene expression was quantified by 2−ΔΔCt. The primers are as follows:
Circ-PRMT5 Forward: GTACCGTTATGGGCTGCTGT
Reverse: TATGCATTTCCCTCCCTCTG
miR-145 Forward: 5′-CCCGCCGTCCAGTTTTCC-3′
Reverse: 5′-CCGTCGGTGAGGGATTCCT-3′
miR-1304 Forward: GTCGTATCCAGTGCGTGTCG
Reverse: AGAGTGACATCGGAGCTTGGGG
Myc Forward: GGCTCCTGGCAAAAGGTCA
Reverse: CTGCGTAGTTGTGCTGATGT
U6 Forward: 5′-TGCTTCGGCAGCACATATAC-3′
Reverse: 5′-AGGGGCCATGCTAATCTTCT-3′
GAPDH Forward: 5′-TGCACCACCAACTGCTTAGC-3′
Reverse: 5′-GGCATGGACTGTGGTCATGAG-3′
Lentiviral vector, siRNA and mimics
GC cells (MKN-28 and AGS) were infected with the lentivirus with circ-PRMT5 overexpression vector, the mimics or siRNA, and the stable cells were selected with puromycin.
Cell proliferation assay
Cell proliferation was detected using a CCK-8 assay kit. Cells were seeded in a 96-well plate (3000 cells/well). Then, 10 μL of CCK-8 reagent was directly added into the culture medium on the indicated time to measure the optional density (OD) at 450 nm after incubation for 2.5 h at 37 °C.
Colony formation assay
Cells were plated in a 6-well plate (1 × 103 cells/well) and incubated for 7 d at 37 °C with medium change every 2 d. On day 7, cells were fixed in 4% paraformaldehyde and stained with a crystal violet solution. Cell colonies were then counted and analyzed.
Transwell migration and invasion assay
The 24-well Transwell (Corning Costar, Corning, NY, 8.0 μm pore size) insert chamber plates were used for cell migration and invasion assay without (for migration assay) or with (for invasion assay) coated Matrigel in the upper surfaces of the transwell filters. Cells (3 × 105) in 200 μL of serum-free medium were loaded to the upper chamber. A total of 500 μL of medium with 10% FBS was added into the lower chamber. After 48 h incubation, the non-migrated and non-invaded cells on the upper side of the chamber were scraped off with a cotton swab, and the cells migrated or invaded to the opposite side were fixed in 4% paraformaldehyde and stained with a crystal violet solution. The cell numbers were then counted and analyzed.
Flow cytometric analysis
To identify cell apoptosis, digested GC cells were washed with cold PBS and re-suspended in binding buffer and fixed, then incubated with APC-AnnexinV and 7-AAD at room temperature for 15 min in the dark. Then, the fluorescence was measured with a BD FACSCalibur flow cytometer (BD) after Annexin V binding buffer was added to the mixture. Cell Quest software (Becton Dickinson, Franklin Lakes, NJ) was used to investigate the cell apoptosis.
Luciferase reporter assay
WT or MUT plasmid of circ-PRMT5 or MYC-3′UTR were co-infected with miR-145/miR-145 mimics into GC cells seeded in a 96-well plate. The miR-NC was used as negative control. The cells were collected after 48 h, the Firefly and Renilla luciferase activities were determined with a dual-luciferase reporter assay system. The relative luciferase activity was normalized to Renilla luciferase activity.
Cytoplasmic and nuclear RNA extraction
Cytoplasmic and Nuclear RNAs were extracted using the RNA subcellular isolation kit following the manufacturer’s instructions.
Western blotting analysis
Total proteins from CG cells were purified using RIPA lysis buffer. Equal amounts of proteins were separated on 12% SDS-PAGE gels and transferred onto PVDF membranes. The primary antibodies were diluted 1:1000 according to the instructions and incubated overnight at 4 °C. HRP rabbit IgG secondary antibodies were incubated at a dilution of 1:2000 at room temperature for 1 h. The membranes were washed three times with TBST and visualized using SuperSignal West Dura Extended Duration Substrate following the manufacturer’s instructions.
Statistical analysis
Statistical analyses were performed using SPSS 20.0 (IBM, SPSS, Chicago, IL) and GraphPad Prism (GraphPad, La Jolla, CA). Student’s t-test or chi-square test was used to evaluate the statistical significance for two group comparisons. OS curves were evaluated with the Kaplan–Meier method. Pearson correlation analysis was used for the association between genes. p < .05 was statistically significant.
Results
High expression of circ-PRMT5 indicated poor prognosis in GC patients
To investigate the clinical significance of circ-PRMT5 expression in GC patients, we collected 90 pairs of GC and self-matched adjacent normal tissues from the GC patients. The circ-PRMT5 expression in these 90 GC patients was measured by qRT-PCR. Compared with that in adjacent normal tissues, circ-PRMT5 expression was significantly increased in GC tissues (, p < .001). We further confirmed this result by comparing the circ-PRMT5 expression between the GC cells and the normal human gastric epithelial cells. As shown in , circ-PRMT5 was dramatically higher in all tested GC cells (AGS, MKN-28, MKN-45, BGC-823, MGC803, SGC-7901) than that in the normal human gastric epithelial GES-1 cells (p < .001).
Figure 1. Upregulaed circ-PRMT5 was identified in GC tissues and acted as an independent prognostic factor for poor OS in GC patients. (A) circ-PRMT5 expression was statistically significantly higher in GC tissues than in paired normal adjacent tissues from 90 GC patients tested by qRT-PCR; (B) circ-PRMT5 expression was statistically significantly higher in the GC cells (AGS, MKN-28, MKN-45, BGC-823, MGC803, SGC-7901) than in the normal human gastric epithelial GES-1 cells tested by qRT-PCR (p < .05); (C) Kaplan–Meier analysis of the association between the circ-PRMT5 expression levels with the OS of GC patients. The 90 patients were divided into circ-PRMT5 low expression group (n = 45) and circ-PRMT5 high expression group (n = 45) based on the median expression value of circ-PRMT5 in GC tissues, then the Kaplan–Meier survival curves were used to compare the OS between the two groups. *p < .05; **p < .01; ***p < .001.
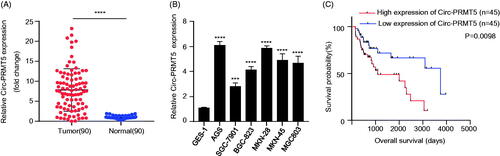
Next, we analyzed whether circ-PRMT5 acts as an independent prognostic factor for GC patients. The GC patients were divided into two groups, circ-PRMT5 high expression group and circ-PRMT5 low expression group, using the median circ-PRMT5 expression level in GC1 tissues as the cutoff value. The association between the circ-PRMT5 expression and the clinicalpathological parameters of GC patients in circ-PRMT5 low expression group (n = 45) and circ-PRMT5 high expression group (n = 45) was analyzed by the chi-square test. We found that the high circ-PRMT5 expression was closely related with the tumour size, the TNM stages, the degree of differentiation, the lymph node metastasis and the distant metastasis regardless of the patient’s age and gender, and the difference was statistically significant (, p < .001).
Table 1. Association between circ-PRMT5 expression and GC clinicopathological characteristics in GC patients were analyzed by chi-square test.
In addition, further statistical analyses with the Kaplan–Meier method showed that GC patients with low circ-PRMT5 expression had longer overall survival (OS) times than those with high circ-PRMT5 expression, which indicated that high circ-PRMT5 expression in GC patients was negatively associated with patient prognosis and displayed a poorer OS (, p < .001).
Circ-PRMT5 induced proliferation and inhibited apoptosis in GC cells
To determine the biological roles of circ-PRMT5 in GC cells, we first designed two siRNAs targeting circ-PRMT5 (designated as si-Circ-PRMT5#1 and si-Circ-PRMT5#2, respectively) and infected into the two GC cell lines (AGS and MKN-28) with the highest circ-PRMT5 expression level to silence the circ-PRMT5 expression. The results showed that infection of si-Circ-PRMT5#1 or si-Circ-PRMT5#2 resulted in a statistically significant decrease in circ-PRMT5 expression level (p < .01); compared with si-NC-PRMT5, both si-Circ-PRMT5#1 and si-Circ-PRMT5#2 could knock down more than 50% of circ-PRMT5 expression in both AGS and MKN-28 cells detected by qRT-PCR assay (. Furthermore, the CCK-8 assay was used to detect the cell viability of AGS and MKN-28 cells (si-NC, si-Circ-PRMT5#1, si-Circ-PRMT5#2) at 0, 24, 48, 72, and 96 h, our results showed that silence of circ-PRMT5 statistically significantly inhibited the viability (OD value) of both AGS (, left panel) and MKN-28 (, right panel) cells at 450 nm (p < .001), si-Circ-PRMT5#1 and si-Circ-PRMT5#2 showed almost the same efficacy; clone formation assay indicated that silence of circ-PRMT5 statistically significantly inhibited the clone formation ability (p < .001), the images (, left panel) and numbers (, middle and right panels) of the clones were acquired under the microscope; meanwhile, flow cytometric assay showed that the percentage of apoptosis was statistically significantly increased in the AGS (p < .01) and MKN-28 (p < .05) cells after circ-PRMT5 knockdown (.
Figure 2. Circ-PRMT5 promoted viability, colony formation, migration, invasion and inhibited apoptosis in GC cells. (A) qRT-PCR analysis confirmed that circ-PRMT5 was successfully silenced with si-circPRMT5#1 or si-circPRMT5#2 containing lentivirus infection; the following indicators were then assessed in the AGS and MKN-45 cells infected with the si-NC, si-CircPRMT5#1 or si-CircPRMT5#2 containing lentivirus: (B) cell viability, (C) clone formation assays, (D) cell apoptosis, (E) cell migration, and (F) cell invasion. *p < .05; **p < .01; ***p < .001.
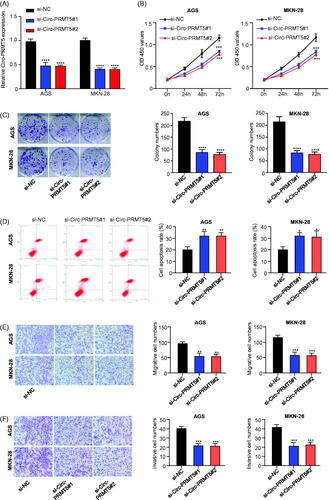
Circ-PRMT5 promoted EMT in GC cells
It is known that EMT is a critical process that induces aggressiveness in cancer cells. Therefore, the transwell migration and invasion assays were used to assess if circ-PRMT5 could enhance EMT phenotypes in CG cells. Our results showed that silence of circ-PRMT5 statistically significantly inhibited the migration () and invasion () abilities of the GC cells, the images (, left panel) and numbers of the migrated (, middle and right panels) and invaded (, middle and right panels) cells were acquired under the microscope.
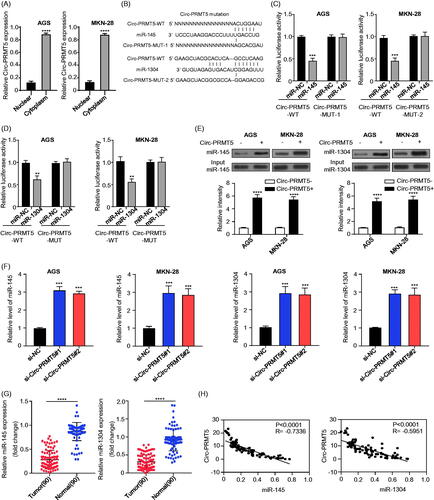
Circ-PRMT5 was confirmed to sponge miR-145 and miR-1304 in GC cells
As previously reported, circRNAs primarily function as miRNA sponges to regulate gene expression and our mRNA fractionation examination revealed that the majority of circ-PRMT5 localized in the cytoplasm (. Therefore, we examined the potential miRNAs associated with circ-PRMT5. CircInteractome was used to predict the potential target miRNAs that could bind with the circ-PRMT5 sequence, and miR-145 and miR-1304 were selected as the best potential targets of circ-PRMT5 (. To investigate whether miR-145 and miR-1304 can interact with circ-PRMT5 in GC cells, we first performed luciferase reporter gene assay in AGS and MKN-28 cells, respectively, our results showed that overexpression of miR-145 or miR-1304 inhibited luciferase activity of the wild-type circ-PRMT5 reporter gene in AGS (, left panel) and MKN-28 (, right panel) cells, while this decreased luciferase activity of circPRMT5 reporter gene was disappeared when the predicted binding sites of circ-PRMT5 with miR-145 or miR-1304 were mutated (), these results suggested that circ-PRMT5 may function as a sponge to miR-145 and miR-1304 in GC cells. To further confirm this result, the interact between circ-PRMT5 with miR-145 or miR-1304 was detected by RNA in vivo precipitation (RIP), it was confirmed that the circ-PRMT5 probe could pull down more miR-145/miR-1304 than the control oligo probe in AGS and MKN-28 cells, the difference was statistically significant (, p < .001); furthermore, the functional association between circ-PRMT5 with the miR-145 and miR-1304 was investigated by measure of miR-145 and miR-1304 expression in circ-PRMT5 silenced AGS and MKN-28 cells, and the GC and paired adjacent normal gastric tissues from 90 GC patients, the results showed that miR-145 and miR-1304 expression was statistically significantly upregulated when circ-PRMT5 was silenced in AGS and MKN-28 cells (, p < .001); the expression of miR-145 and miR-1304 was statistically significantly lower in GC tissues than in the paired adjacent normal gastric tissues (, p < .001). Meanwhile, the Pearson correlation coefficient was used to analyze the correlation between circ-PRMT5 and miR-145/miR-1304 expression, which showed that circ-PRMT5 expression was significantly negatively correlated with miR-145 (, left panel) or miR-1304 (, right panel) expression (p < .01). These results suggested that circ-PRMT5 directly interacted with miR-145/miR-1304, resulting in the aggressiveness of the GC cells.
Figure 3. Identification of the potential miRNAs sponged by circ-PRMT5. (A) Sublocalization of circ-PRMT5 was determined by detecting the circ-PRMT5 expression levels in the cytoplasmic and nuclear RNA extraction from AGS and MKN-28 cells, U6 and GAPDH are used as the internal reference in the nucleus and cytoplasm, respectively; (B) Schematic representation of the potential binding sites of miR-145 and miR-1304 with circ-PRMT5 (https://circinteractome.nia.nih.gov/index.html); (C) double luciferase reporter assay in the AGS and MKN-28 cells co-infected with circ-PRMT5 WT or mutated reporter and miR-145 mimics; (D) double luciferase reporter assay in the AGS and MKN-28 cells co-infected with circ-PRMT5 WT or mutated reporter and miR-1304 mimics; (E) Circ-PRMT5 in GC cell lysates was pulled down and enriched with biotin-labelled circ-PRMT5-specific probe; (F) the expression levels of miR-145 and miR-1304 in circ-PRMT5 silenced AGS and MKN-28 cells detected by qRT-PCR; (G) the expression levels of miR-145 and miR-1304 in paired GC and adjacent normal tissues from 90 GC patients (same samples as in ) detected by qRT-PCR; (H) Pearson correlation analysis of the association between miR-145/miR-1304 with circ-PRMT5.
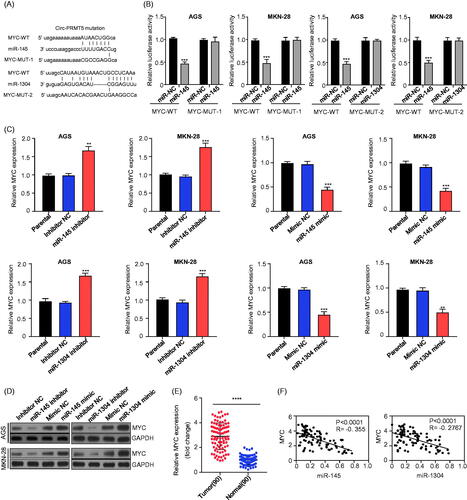
MYC was validated as a target gene of miR-145 and miR-1304 in GC cells
Next, we used miRanda (http://www.microrna.org/) to predict the target gene of miR-145 and miR-1304 in GC cells; and miR-145 and miR-1304 binding sites were found in the 3′ non-coding region of the classical oncogene MYC, thus showed the best potential (. GC cells co-infected with plasmid containing 3′-UTR-WT regions of MYC with miR-145/miR-1304 mimics had significantly less relative luciferase activity than the controls (miR-NC), while mutation of the potential miR-145/miR-1304 binding sites in the MYC 3′-UTR abolished this effect (, p < .001), these results suggested that MYC could be a putative target of miR-145/miR-1304. Furthermore, the functional association between MYC with the miR-145 and miR-1304 was investigated by measure of the mRNA and protein expression levels of MYC after knockdown (inhibitor)/overexpression(mimic) of miR-145 or miR-1304 in AGS and MKN-28 cells, and in the GC and paired adjacent normal gastric tissues from 90 GC patients, the results showed that knockdown/overexpression of miR-145 or miR-1304 statistically significant increased/decreased MYC mRNA (, p < .001) and protein () expression, respectively; the mRNA expression levels of MYC in GC tissues was statistically significantly increased (, p < .001). Meanwhile, Pearson correlation coefficient was used to analyze the correlation between MYC and miR-145/miR-1304 expression, which showed that MYC expression was significantly negatively correlated with miR-145 (, left panel) or miR-1304 (, right panel) expression (p < .0001). These results suggested that MYC could directly interact with miR-145/miR-1304 to promote the development of GC.
Figure 4. MYC was the downstream target of miR-145 and miR-1304 in GC cells. (A) Schematic representation of the potential binding sites of miR-145 and miR-1304 with MYC-3′UTR (http://www.microrna.org/); (B) double luciferase reporter assay in the AGS and MKN-28 cells infected with WT or mutated MYC-3′UTR reporter and miR-145 or miR-1304 mimics; (C) the expression levels of MYC mRNA in AGS and MKN-28 cells after knockdown (inhibitor)/overexpression (mimic) of miR-145 or miR-1304 detected by qRT-PCR; (D) the protein expression levels of MYC in AGS and MKN-28 cells after knockdown (inhibitor)/overexpression (mimic) of miR-145 or miR-1304 detected by Western-Blot; (E)The expression levels of MYC in paired GC and adjacent normal tissues from 90 patients (same samples as in ) detected by qRT-PCR; (F) Pearson correlation analysis of the association between miR-145/miR-1304 with MYC.
Circ-PRMT5 sponged miR-145 and miR-1304 in GC cells to upregulate MYC
To determine whether circ-PRMT5 sponges miR-145 and miR-1304 to regulate MYC expression, we first measured MYC expression levels by qRT-PCR and Western Blotting. Our results found that circ-PRMT5 silence considerably downregulated MYC expression at both the mRNA and protein levels in GC cells, and these effects could be partially reversed by co-infection of the miR-145 or miR-1304 inhibitor (, p < .001). Pearson correlation analysis of the association between circ-PRMT5 and MYC expression in GC tissues from 90 GC patients (same samples as in ) showed that circ-PRMT5 expression was statistically significantly and positively correlated with MYC expression (, p < .0001). Cell viability in AGS and MKN-28 cells (infected with si-NC, si-Circ-PRMT5#1, si-Circ-PRMT5#1 + miR-145 inhibitor, si-Circ-PRMT5#1 + miR-1304 inhibitor or si-Circ-PRMT5#1 + MYC) at different time points were detected by CCK-8 assay (), our results showed that knockdown of circ-PRMT5 reduced the OD value at 450 nm, and this effects could be partially reversed by co-infection of the miR-145/miR-1304 inhibitor or MYC overexpression vector (, p < .001).
Figure 5. Circ-PRMT5 promoted GC progression by regulation of miR-145/1304/MYC axis. (A) The expression levels of MYC mRNA in AGS and MKN-28 cells with the infection of si-NC, si-circPRMT5#1, si-circPRMT5#1 + miR-145 inhibitor or si-circ-PRMT5#1 + miR-1304 inhibitor detected by qRT-PCR; (B) the expression levels of MYC protein in AGS and MKN-28 cells with the infection of si-NC, si-circ-PRMT5#1, si-circ-PRMT5#1 + miR-145 inhibitor or si-circ-PRMT5#1 + miR-1304 inhibitor detected by Western-Blotting; (C) Pearson correlation analysis of the association between circ-PRMT5 with MYC in GC tissues from 90 patients (same samples as in ); (D) cell viability in AGS and MKN-28 cells with the infection of si-NC, si-circ-PRMT5#1, si-circ-PRMT5#1 + miR-145 inhibitor, si-circ-PRMT5#1 + miR-1304 inhibitor or si-circ-PRMT5#1 + MYC, at different time points, detected by CCK-8 assay; (E) Clone formation assay in AGS and MKN-28 cells with the infection of si-NC, si-circ-PRMT5#1, si-circ-PRMT5#1 + miR-145 inhibitor, si-circ-PRMT5#1 + miR-1304 inhibitor or si-circ-PRMT5#1 + MYC; (F) cell migration ability in AGS and MKN-28 cells with the infection of si-NC, si-circ-PRMT5#1, si-circ-PRMT5#1 + miR-145 inhibitor, si-circ-PRMT5#1 + miR-1304 inhibitor or si-circ-PRMT5#1 + MYC detected by Transwell assay; (G) cell invasion ability in AGS and MKN-28 cells with the infection of si-NC, si-circ-PRMT5#1, si-circ-PRMT5#1 + miR-145 inhibitor, si-circ-PRMT5#1 + miR-1304 inhibitor or si-circ-PRMT5#1 + MYC detected by Transwell with matrigel.
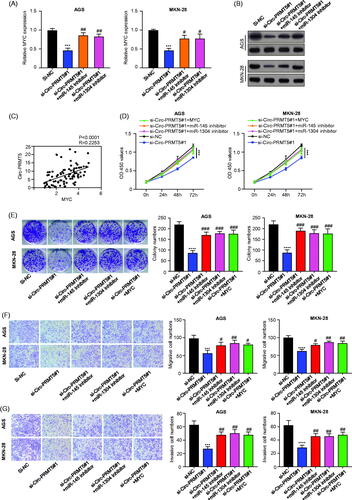
Clone formation assay in AGS and MKN-28 cells (infected with si-NC, si-circ-PRMT5#1, si-circ-PRMT5#1 + miR-145 inhibitor, si-circ-PRMT5#1 + miR-1304 inhibitor or si-circ-PRMT5#1 + MYC) was performed, our results showed that knockdown of circ-PRMT5 reduced the clone formation ability, and this effects could be partially reversed by co-infection of the miR-145/miR-1304 inhibitor or MYC overexpression vector (, p < .001). Cell migration ability in AGS and MKN-28 cells (infected with si-NC, si-circ-PRMT5#1, si-circ-PRMT5#1 + miR-145 inhibitor, si-circ-PRMT5#1 + miR-1304 inhibitor or si-circ-PRMT5#1 + MYC) was detected by Transwell assay, our results showed that knockdown of circ-PRMT5 reduced the migration ability, and this effects could be partially reversed by co-infection of the miR-145/miR-1304 inhibitor or MYC overexpression vector (, p < .001). Cell invasion ability in AGS and MKN-28 cells (infected with si-NC, si-circ-PRMT5#1, si-circ-PRMT5#1 + miR-145 inhibitor, si-circ-PRMT5#1 + miR-1304 inhibitor or si-circ-PRMT5#1 + MYC) was detected by Transwell with matrigel, our results showed that knockdown of circ-PRMT5 reduced the invasion ability, and this effects could be partially reversed by co-infection of the miR-145/miR-1304 inhibitor or MYC overexpression vector (, p < .001).
Discussion
CircRNAs are a new group of ncRNAs with fundamentally unclear biological functions. Evidences demonstrated that circRNAs are rich in cells and show cell/tissue-specific expression, suggesting circRNAs may be vital in regulating gene transcription and expression, thus the essential functions of circRNAs in various biological and pathological procedures [Citation19]. In recent years, researchers have focused extensive attention on circRNAs due to their important functions in many cellular mechanisms.
CircRNAs also have the potential to be as encouraging biomarkers for diagnosis of many diseases such as cancer [Citation20]. Therefore, the circRNA function in carcinogenesis has recently attracted researcher’s attention. The gene expression regulation varies of CircRNAs in different types or different stages of cancer [Citation19]. CircRNAs are well known to act as miRNA sponges to form the circRNA–miRNA–mRNA axis [Citation8,Citation13,Citation14,Citation16,Citation21–23].
Circ-PRMT5 is a newly identified circRNA, plays critical role in promoting cell EMT of urothelial carcinoma of the bladder (UCB) and may serve as an exploitable therapeutic target for UCB patients. However, the expression and function of circ-PRMT5 in GC remain unknown.
Here, we investigated the GC tissues and paired adjacent normal tissues from 90 GC patients and found that circ-PRMT5 expression was significantly higher in the GC tissues than in the matched adjacent normal tissues. Increased circ-PRMT5 expression was significantly correlated with the poor prognosis in GC patients. GC patients with low circ-PRMT5 expression had longer survival times than those with high circ-PRMT5 expression. Therefore, it was illustrated that circ-PRMT5 could serve as a biomarker to predict the survival in GC patients.
MiRNA, a key component of the ncRNA family with the length of less than 22 nucleotides, plays multifaceted roles in controlling cellular functions by degradation of the target genes. Large evidences have demonstrated that miRNA mediated growth, oxidative response, signalling transduction and cell apoptosis.
In this study, CircInteractome was used to predict the potential miRNAs binding to circ-PRMT5, and miR-145 and miR-1304 were found to be the best prospective candidates. MiR-145 has been reported as a tumour suppressor in many cancer types, such as colorectal cancer [Citation24], cervical carcinoma [Citation25], prostate cancer [Citation26], oesophageal squamous cell carcinoma [Citation27] and gastric cancer [Citation28]. MiR-1304 has also been reported being sponged by circRNAs to promote cancer progression [Citation29,Citation30]. However, the possible mechanism might need additional exploration. In current study, we also found that miR-145 and miR-1304 were down-regulated in the GC tissues and acted as an oncosuppressor in GC patents. MiR-145 and miR-1304 expression was statistically significantly up-regulated when circ-PRMT5 was silenced in AGS and MKN-28 cells; overexpression of miR-145 or miR-1304 inhibited luciferase activity of the wild-type circ-PRMT5 reporter gene in AGS and MKN-28 cells, while this effect was disappeared when the predicted binding sites of circ-PRMT5 were mutated; circ-PRMT5 probe could pull down more miR-145/miR-1304 than the oligo probe; Pearson correlation coefficient analysis showed a negative correlation between the circ-PRMT5 and the miR-145/miR-1304 expression.
MYC is an oncogene involved in cell cycle regulation, cell adhesion, cell growth arrest, metabolism, protein synthesis, ribosome biogenesis and mitochondrial function, which has been described as a crucial element of numerous human carcinogenesis processes, such as GC [Citation31–34]. MiRanda database showed binding sites of miR-145 and miR-1304 in the 3′ non-coding region of MYC. Our further investigate showed that mutation of the potential miR-145/miR-1304 binding sites in the MYC 3′-UTR abolished MYC induced luciferase activity decrease; furthermore, knockdown/overexpression of miR-145 or miR-1304 increased/decreased MYC mRNA and protein expressions; the mRNA expression levels of MYC in GC tissues was also increased; Pearson correlation coefficient analysis showed a negative correlation between the miR-145/miR-1304 and the MYC expression, suggesting MYC could directly interacted with miR-145/miR-1304 to promote the progress of GC.
In summary, our findings revealed that circ-PRMT5 expression was significantly increased in GC patients and cells. High circ-PRMT5 expression was associated with a poor prognosis in GC patients. Functionally, circ-PRMT5 promoted GC cell growth, clone formation, migration and invasion, while inhibited apoptosis by sponging miR-145 and miR-1304 in GC cells to upregulate MYC expression, thus contributed to the GC tumourigenesis. Therefore, circ-PRMT5 may be used as a potential prognostic predictor and therapeutic target in GC patients.
Disclosure statement
The authors declare that they have no conflict of interests for conducting this research.
References
- Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108.
- Allemani C, Weir HK, Carreira H, et al. Global surveillance of cancer survival 1995–2009: analysis of individual data for 25,676,887 patients from 279 population-based registries in 67 countries (CONCORD-2). Lancet. 2015;385(9972):977–1010.
- Shen F, Liu P, Xu Z, et al. CircRNA_001569 promotes cell proliferation through absorbing miR-145 in gastric cancer. J Biochem. 2019;165(1):27–36.
- Wang J, Song YX, Wang ZN. Non-coding RNAs in gastric cancer. Gene. 2015;560(1):1–8.
- Song JH, Meltzer SJ. MicroRNAs in pathogenesis, diagnosis, and treatment of gastroesophageal cancers. Gastroenterology. 2012;143(1):35.e2–47.e2.
- Salzman J, Chen RE, Olsen MN, et al. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9(9):e1003777.
- Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19(2):141–157.
- Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338.
- Cortes-Lopez M, Miura P. Emerging functions of circular RNAs. Yale J Biol Med. 2016;89(4):527–537.
- Barrett SP, Salzman J. Circular RNAs: analysis, expression and potential functions. Development. 2016;143(11):1838–1847.
- Li Z, Huang C, Bao C, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22(3):256–264.
- Chen Y, Li C, Tan C, et al. Circular RNAs: a new frontier in the study of human diseases. J Med Genet. 2016;53(6):359–365.
- Han D, Li J, Wang H, et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66(4):1151–1164.
- Hsiao K-Y, Lin Y-C, Gupta SK, et al. Noncoding effects of circular RNA CCDC66 promote colon cancer growth and metastasis. Cancer Res. 2017;77(9):2339–2350.
- Chen L, Zhang S, Wu J, et al. circRNA_100290 plays a role in oral cancer by functioning as a sponge of the miR-29 family. Oncogene. 2017;36(32):4551–4561.
- Zhong Z, Huang M, Lv M, et al. Circular RNA MYLK as a competing endogenous RNA promotes bladder cancer progression through modulating VEGFA/VEGFR2 signaling pathway. Cancer Lett. 2017;403:305–317.
- Li F, Zhang L, Li W, et al. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/beta-catenin pathway. Oncotarget. 2015;6(8):6001–6013.
- Chen X, Chen R-X, Wei W-S, et al. PRMT5 circular RNA promotes metastasis of urothelial carcinoma of the bladder through sponging miR-30c to induce epithelial-mesenchymal transition. Clin Cancer Res. 2018;24(24):6319–6330.
- Du WW, Yang W, Liu E, et al. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44(6):2846–2858.
- Chaichian S, Shafabakhsh R, Mirhashemi SM, et al. Circular RNAs: a novel biomarker for cervical cancer. J Cell Physiol. 2019;234:11391–11400.
- Hansen TB, Kjems J, Damgaard CK. Circular RNA and miR-7 in cancer. Cancer Res. 2013;73(18):5609–5612.
- Zhang J, Liu H, Hou L, et al. Circular RNA_LARP4 inhibits cell proliferation and invasion of gastric cancer by sponging miR-424-5p and regulating LATS1 expression. Mol Cancer. 2017;16(1):151.
- Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388.
- Reimondez-Troitino S, González-Aramundiz JV, Ruiz-Bañobre J, et al. Versatile protamine nanocapsules to restore miR-145 levels and interfere tumor growth in colorectal cancer cells. Eur J Pharm Biopharm. 2019;140:78–90.
- Ma L, Li LL. miR-145 contributes to the progression of cervical carcinoma by directly regulating FSCN1. Cell Transplant. 2019;28(9-10):1299–1305
- He J-H, Han Z-P, Zou M-X, et al. CDX2/mir-145-5p/SENP1 pathways affect LNCaP cells invasion and migration. Front Oncol. 2019;9:477.
- Zhang Y, Zhang L, Wang R, et al. LncRNA Erbb4-IR promotes esophageal squamous cell carcinoma (ESCC) by downregulating miR-145. J Cell Biochem. 2019;120(10):17566-17572.
- Obermannova R, Redova-Lojova M, Vychytilova-Faltejskova P, et al. Tumor expression of miR-10b, miR-21, miR-143 and miR-145 is related to clinicopathological features of gastric cancer in a central European population. Anticancer Res. 2018;38(6):3719–3724.
- Liu G, Shi H, Deng L, et al. Circular RNA circ-FOXM1 facilitates cell progression as ceRNA to target PPDPF and MACC1 by sponging miR-1304-5p in non-small cell lung cancer. Biochem Biophys Res Commun. 2019;513(1):207–212.
- Pan Y, Xu T, Liu Y, et al. Upregulated circular RNA circ_0025033 promotes papillary thyroid cancer cell proliferation and invasion via sponging miR-1231 and miR-1304. Biochem Biophys Res Commun. 2019;510(2):334–338.
- Oster SK, Ho CSW, Soucie EL, et al. The myc oncogene: MarvelouslY complex. Adv Cancer Res. 2002;84:81–154.
- Cole MD, Henriksson M. 25 years of the c-Myc oncogene. Semin Cancer Biol. 2006;16(4):241.
- Vita M, Henriksson M. The Myc oncoprotein as a therapeutic target for human cancer. Semin Cancer Biol. 2006;16(4):318–330.
- Zhang C, Wang W, Lin J, et al. lncRNA CCAT1 promotes bladder cancer cell proliferation, migration and invasion. Int Braz J Urol. 2019;45(3):549–559.
