Abstract
LncRNA LINC01518 was previously reported to be upregulated in oesophageal squamous cell carcinoma (ESCC) tissues compared with normal tissues. However, there are no previous studies concerning the specific function and molecular mechanism of LINC01518 in ESCC. LINC01518 and miR-1-3p expression levels in ESCC cells were detected by qRT-PCR. Cell proliferation and apoptosis were evaluated by CCK-8 and flow cytometry analysis, respectively. The relationships between LINC01518, phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit α (PIK3CA) and miR-1-3p were explored by luciferase reporter assay. The alteration of the PIK3CA/protein kinase B (Akt) pathway was examined by Western blot. We found that LINC01518 expression was upregulated and miR-1-3p expression was downregulated in ESCC cells. LINC01518 knockdown inhibited cell proliferation and promoted apoptosis in ESCC cells. In addition, LINC01518 functioned as a competing endogenous RNA (ceRNA) for miR-1-3p in ESCC cells and miR-1-3p downregulation blocked the effects of LINC01518 knockdown on cell proliferation and apoptosis in ESCC cells. Moreover, PIK3CA was identified as a target of miR-1-3p and LINC01518 knockdown inhibited the PIK3CA/Akt pathway by upregulating miR-1-3p in ESCC cells. In conclusion, LINC01518 knockdown inhibited tumorigenicity in ESCC cells by suppression of PIK3CA/Akt pathway through upregulating miR-1-3p.
Introduction
As a highly lethal malignancy, oesophageal cancer is one of the most frequently diagnosed digestive system malignancies with high mortality and morbidity, ranking the sixth in terms of cancer-associated mortality all over the world [Citation1,Citation2]. Oesophageal squamous cell carcinoma (ESCC) is considered as the most common histological subtype of oesophageal cancer in China, accounting for more than 90% of the total oesophageal cancer cases [Citation3]. Majority of the patients with ESCC are often diagnosed at advanced stages due to lack of specific clinical symptoms in early stage [Citation4]. In spite of marked improvements in diagnosis and therapeutic modalities of ESCC recently, the prognosis of patients with ESCC is still unfavourable with an overall 5-year survival rate of less than 20% owing to high recurrence and distant metastasis [Citation5]. Therefore, it is desperately needed to discover the genetic mechanisms underlying ESCC initiation and progression and develop novel effective therapeutic schemes for ESCC.
It has been estimated that only 2% of the human genome encodes proteins while majority of the transcribed RNAs are noncoding RNAs (ncRNAs), which consists of small ncRNAs and long ncRNAs (lncRNAs) [Citation6]. LncRNAs, which have become a focus of the study of ncRNAs, are commonly defined as a group of RNA transcripts greater than 200 nucleotides in length with no or limited protein-coding capacity [Citation7]. There is striking evidence that lncRNAs are recognized as important players in diverse pathophysiological processes, especially tumour progression and serve as tissue-specific oncogenes, tumour suppressor genes or both, depending on the circumstance [Citation8]. An increasing number of studies have shown the involvement of aberrantly expressed lncRNAs in the initiation and progression of various cancers, including ESCC [Citation4]. As a novel member of lncRNAs, LINC01518 was previously reported to be upregulated in ESCC tissues compared with that in normal tissues [Citation9]. However, up to date, there are no studies concerning the specific function and molecular mechanism of LINC01518 in ESCC.
In our study, we determined the expression of LINC01518 in ESCC cells and the effects of LINC01518 on ESCC cell proliferation and apoptosis. In addition, mechanistic analysis found that LINC01518 knockdown inhibited the phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit α (PIK3CA)/protein kinase B (Akt) pathway by competing for microRNA (miRNA)-1, thus inhibiting cell proliferation and promoting apoptosis in ESCC cells.
Materials and methods
Cell culture
The ESCC cell lines (Ec9706, Eca109, TE-1 and KYSE140) and the normal human oesophageal epithelial cell line (Het-1A) were purchased from the American Type Culture Collection (Manassas, VA, USA). All cells were maintained in RPMI-1640 medium (Gibco, Grand Island, NY, USA) supplemented with 10% foetal bovine serum (HyClone, Logan, UT, USA) and 100 U/mL penicillin-streptomycin mixture (Thermo Fisher Scientific, Waltham, MA, USA) in a humidified incubator containing 5% CO2 at 37 °C.
Cell transfection
Small interfering RNA (siRNA) against LINC01518 (si-LINC01518) and siRNA control (si-ctrl), miR-1-3p agomir and agomir control (miR-ctrl), miR-1-3p antagomir (anti-miR-1-3p) and antagomir control (anti-miR-ctrl) were purchased from Shanghai GenePharma Co., Ltd. (Shanghai, China). Ec9706 and TE-1 cells were seeded into 96-well plates and transient transfection was then performed using Lipofectamine 3000 Transfection Reagent (Invitrogen, Carlsbad, CA, USA). Cells were collected for the next experiments at 48 h post-transfection.
RNA extraction and quantitative real-time PCR (qRT-PCR)
Total RNA was isolated from cultured cells using TRIzol Reagent (Takara, Dalian, China) and the concentration of extracted RNA was quantified using a NanoDrop 2000/2000c (Thermo Fisher Scientific). A total of 10 ng of RNA was reversely transcribed into cDNA using RevertAid First Strand cDNA kit (Thermo Fisher Scientific). For the detection of LINC01518 and miR-1-3p expression, qRT-PCR was carried out using a SYBR Green qPCR Mix kit (Takara) and a TaqMan MicroRNA PCR Kit (Applied Biosystems, Foster City, CA, USA), respectively, on the CFX Connect Real-Time System (Bio-Rad, Hercules, CA, USA). The primer sequences were as follows: LINC01518, forward 5′-CAGT GACG GAAC AGTA CCAG-3′ and reverse 5′-TCAC AAAC ATCC CGCT CT-3′. miR-1-3p, forward 5′-CAGT GCGT GTCG TGGA GT–3′ and reverse 5′-GGCC TGGA ATGT AAAG AAGT–3′; GAPDH, forward 5′-GAGT CAAC GGAT TTGG TCGT–3′ and reverse 5′-TTGA TTTT GGAG GGAT CTCG–3′; U6, forward 5′-GCTT CGGC AGCA CATA TACT AAAT–3′ and reverse 5′-CGCT TCAC GAAT TTGC GTGT CAT–3′. The relative gene expression level was compared using the 2−ΔΔCt method with GAPDH and U6 small nuclear RNA (snRNA) as an endogenous control for lncRNA and miRNA, respectively.
Cell proliferation assay
Cell proliferation was detected using Cell Counting Kit-8 assay kit reagent (CCK-8; Dojindo, Kumamoto, Japan). Ec9706 and TE-1 cells were seeded into 96-well plates at 2 × 104 cells/well and cotransfected with si-LINC01518 or si-ctrl and anti-miR-1-3p or anti-miR-ctrl. Following culturing for the indicated time (24, 48, and 72 h), 10 µl of CCK-8 solution was added into each well and cells were incubated for 4 h. The optical density was measured at a wavelength of 450 nm using ELX-800 microplate reader (Bio-Rad).
Flow cytometry for apoptosis analysis
The transfected Ec9706 and TE-1 cells were harvested 48 h following transfection, washed with cold PBS, and resuspended in 1 × binding buffer. After double staining with Annexin V-fluorescein isothiocyanate (FITC) and propidium iodide (PI) using Annexin V-FITC/PI cell apoptosis detection kit (BD Biosciences, San Diego, CA, USA), the apoptotic cells were examined using a flow cytometer (FACScan, BD Biosciences) equipped with CellQuest Pro Software version 5.1 (BD Biosciences).
Luciferase reporter assay
The wild-type or mutant fragments of LINC01518 and PIK3CA 3′UTR containing the putative binding sites for miR-1-3p were amplified by PCR and inserted into pMIR-REPORT luciferase reporter vector (Promega, Madison, WI, USA), namely LINC01518-WT, LINC01518-MUT, PIK3CA-WT, and PIK3CA-MUT, respectively. Ec9706 cells were plated into 96-well plates and cotransfected with miR-1-3p or miR-ctrl and the constructed luciferase reporter plasmids using Lipofectamine 3000 Transfection Reagent (Invitrogen). The relative luciferase activity was detected at 48 h following transfection using the dual-luciferase reporter assay (Promega).
Western blot analysis
Total protein was extracted from cultured cells using RIPA buffer (Sangon Biotech, Shanghai, China) containing protease inhibitors (Roche, Diagnostics GmbH, Mannheim, Germany). Equal quantities of protein extracts (20 μg/lane) were subjected to 10% SDS-PAGE gel and electro-transferred onto nitrocellulose membranes (Sigma, St. Louis, MO, USA). The membranes were blocked in 5% skimmed milk in Tris-buffered saline containing 0.1% Tween-20 for 1 h at room temperature, followed by incubation overnight at 4 °C with the following primary antibodies against PIK3CA (Abcam, Cambridge, MA, USA; 1:500 dilution), phosphorylated Akt (p-Akt) (Abcam; 1:500 dilution), Akt (Abcam; 1:500 dilution) and β-actin (Abcam; 1:1000 dilution). Following rinsing, the membranes were then incubated with corresponding horseradish peroxidase-conjugated mouse anti-rabbit secondary antibody (Abcam; 1:2000 dilution) for 2 h at room temperature. Immunolabelling was visualized by enhanced chemiluminescent (ECL) system (Bio-Rad).
Statistical analysis
All results are shown as mean ± standard deviation of three independent experiments. Statistical analyses were performed using SPSS version 20.0 software (SPSS, Inc., Chicago, IL, USA). Statistical significance was analyzed by student’s t-test or one-way analysis of variance (ANOVA). p Values less than .05 were considered statistically significant.
Results
The expression of LINC01518 and miR-1-3p in ESCC cells
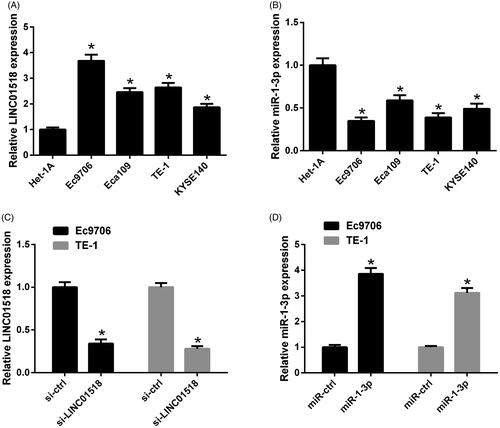
Expression patterns of LINC01518 and miR-1-3p in ESCC cell lines were detected by qRT-PCR. As shown in , LINC01518 expression was upregulated and miR-1-3p expression was downregulated in ESCC cell lines including Ec9706, Eca109, TE-1 and KYSE140 when compared with normal human oesophageal epithelial cell line (Het-1A), especially in Ec9706 and TE-1 cells. Ec9706 and TE-1 cells were, therefore, chosen for further experiments. To understand the functional significances of LINC01518 and miR-1-3p in ESCC cells, Ec9706 and TE-1 cells were transfected with si-LINC01518, miR-1-3p or respective controls. LINC01518 expression was distinctly reduced following transfection with si-LINC01518 () and miR-1-3p expression was dramatically enhanced after introduction with miR-1-3p () in both Ec9706 and TE-1 cells.
Figure 1. Expression profiles of LINC01518 and miR-1-3p in ESCC cells. qRT-PCR analysis of LINC01518 (A) and miR-1-3p (B) in ESCC cell lines (Ec9706, Eca109, TE-1 and KYSE140) and normal human oesophageal epithelial cell line (Het-1A). (C) The expression of LINC01518 in Ec9706 and TE-1 cells after transfection with si-LINC01518 or si-ctrl for 48 h was examined by qRT-PCR. (D) The expression of miR-1-3p in Ec9706 and TE-1 cells was determined by qRT-PCR 48 h after transfection with miR-1-3p or miR-ctrl. *p < .05.
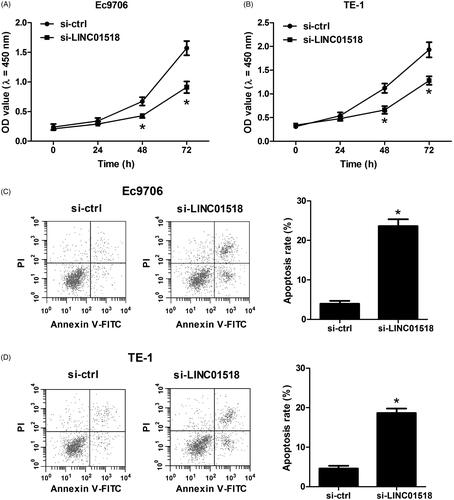
LINC01518 knockdown inhibited proliferation and promoted apoptosis in ESCC cells
The results of CCK-8 assay showed that knockdown of LINC01518 remarkably impeded cell proliferation in Ec9706 () and TE-1 () cells relative to the control group. In addition, flow cytometry assay revealed that the percentage of apoptotic Ec9706 () and TE-1 () cells were substantially increased after decreased expression of LINC01518. These results demonstrated that LINC01518 knockdown inhibited proliferation and promoted apoptosis in ESCC cells.
Figure 2. Effect of LINC01518 knockdown on cell proliferation and apoptosis in ESCC cells. (A and B) Ec9706 and TE-1 cells were transfected with si-LINC01518 or si-ctrl for 0, 24, 48, and 72 h, followed by assessment of cell proliferation by CCK-8 assay. (C and D) Ec9706 and TE-1 cells were transfected with si-LINC01518 or si-ctrl for 48 h, followed by the assessment of apoptosis by flow cytometry analysis. *p < .05.
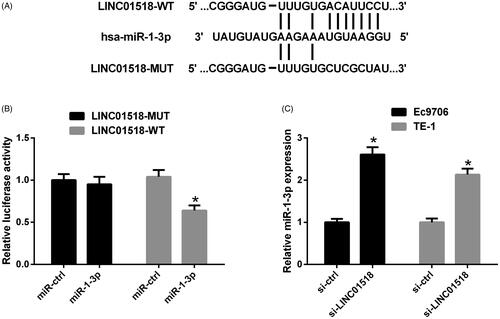
LINC01518 directly targeted miR-1-3p in ESCC cells
Using publicly available algorithms (StarBase and TargetScan), we found that there were potential binding sites between miR-1-3p and LINC01518. To further clarify the relationship between LINC01518 and miR-1-3p, the wild-type and mutated sequences of LINC01518 containing the predicted miR-1-3p binding sites were cloned into pMIR-REPORT luciferase reporter vector and designated as LINC01518-WT and LINC01518-MUT, respectively, as shown in . The subsequent luciferase reporter assay manifested that augmented expression of miR-1-3p markedly reduced the luciferase activity of LINC01518-WT, but showed no significant influence on the luciferase activity of LINC01518-MUT in Ec9706 cells (). To explore the effect of LINC01518 on miR-1-3p expression, expression of miR-1-3p was detected in Ec9706 and TE-1 cells following delivery of si-LINC01518 or si-ctrl. The result of qRT-PCR exhibited that miR-1-3p expression was notably elevated in si-LINC01518-treated Ec9706 and TE-1 cells versus that in si-ctrl-introduced cells (). Altogether, the above results implied that LINC01518 functioned as a ceRNA for miR-1-3p in ESCC cells.
Figure 3. The interaction between LINC01518 and miR-1-3p in ESCC cells. (A) The wild-type and mutated sequences of LINC01518 containing binding sequences complementary to miR-1-3p seed regions. (B) Luciferase activity was measured by luciferase activity assay in Ec9706 and TE-1 cells 48 h after cotransfection with LINC01518-WT or LINC01518-MUT and miR-1-3p or miR-ctrl. (C) miR-1-3p expression was estimated in Ec9706 and TE-1 cells 48 h after transfection with si-LINC01518 or si-ctrl. *p < .05.
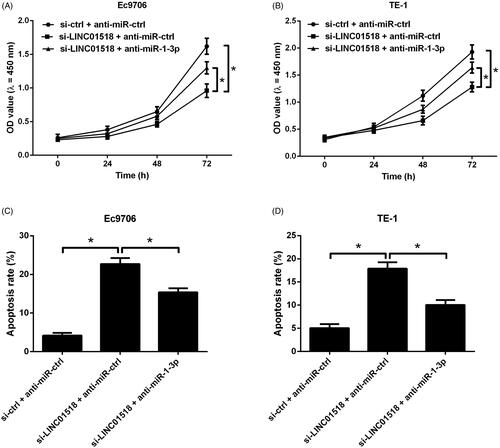
miR-1-3p downregulation blocked the effects of LINC01518 knockdown on cell proliferation and apoptosis in ESCC cells
To investigate whether miR-1-3p inhibition reversed the effects of LINC01518 knockdown on cell proliferation and apoptosis in ESCC cells, Ec9706 and TE-1 cells were cotransfected with si-LINC01518 or si-ctrl and anti-miR-1-3p or anti-miR-ctrl. CCK-8 assay proved that miR-1-3p suppression effectively countervailed the inhibitory effect of LINC01518 silencing on cell proliferation in Ec9706 () and TE-1 () cells. Flow cytometry analysis demonstrated that the depletion of LINC01518-induced apoptosis in Ec9706 () and TE-1 () cells was undermined following reintroduction with anti-miR-1-3p. These results revealed that miR-1-3p inhibition abolished the effects of LINC01518 knockdown on cell proliferation and apoptosis in ESCC cells.
Figure 4. Effects of LINC01518 silencing or combined with miR-1-3p downregulation on cell proliferation and apoptosis in ESCC cells. (A and B) Ec9706 and TE-1 cells were cotransfected with si-LINC01518 or si-ctrl and anti-miR-1-3p or anti-miR-ctrl for 0, 24, 48, and 72 h, followed by the assessment of cell proliferation by CCK-8 assay. (C and D) Ec9706 and TE-1 cells were cotransfected with si-LINC01518 or si-ctrl and anti-miR-1-3p or anti-miR-ctrl for 48 h, followed by the assessment of apoptosis by flow cytometry analysis. *p < .05.
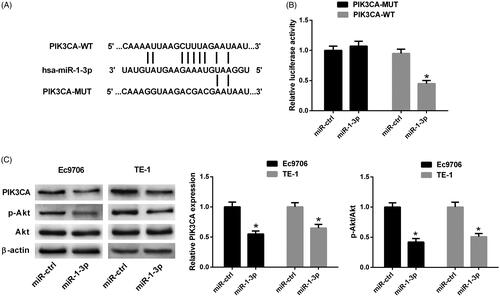
PIK3CA was a target of miR-1-3p in ESCC cells
Through StarBase and TargetScan database prediction, it was predicted that PIK3CA 3′UTR contained putative recognition sites for miR-1-3p. Therefore, luciferase reporter plasmids were constructed incorporating a wild-type or mutated 3′UTR of PIK3CA in which the sequence corresponding to the seed region was altered (). Luciferase reporter assay presented that luciferase activity was dramatically decreased after cotransfection with PIK3CA-WT and miR-1-3p in Ec9706 cells. However, there was no dramatic change in the luciferase activity of Ec9706 cells cotransfected with PIK3CA-MUT and miR-1-3p compared with that transfected with PIK3CA-MUT and miR-ctrl (). The effects of miR-1-3p on the PIK3CA/Akt pathway in ESCC cells were determined by Western blot. As shown in , miR-1-3p overexpression distinctly suppressed the protein levels of PIK3CA and p-Akt in Ec9706 and TE-1 cells with respect to the control group, suggesting that miR-1-3p overexpression inactivated the PIK3CA/Akt pathway in ESCC cells.
Figure 5. The interaction between PIK3CA and miR-1-3p in ESCC cells. (A) Schematic of the wild-type or mutated PIK3CA 3′UTR containing miR-1-3p binding sites. (B) Luciferase activity was measured by luciferase reporter assay 48 h after Ec9706 cells were cotransfected with PIK3CA-WT or PIK3CA-MUT and miR-1-3p or miR-ctrl. (C) The protein levels of PIK3CA, p-Akt and Akt were detected by Western blot in Ec9706 and TE-1 cells 48 h after transfection with miR-1-3p or miR-ctrl. *p < .05.
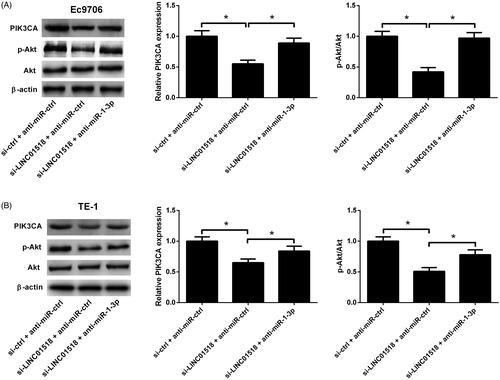
LINC01518 knockdown inhibited PIK3CA/Akt pathway in ESCC cells by regulating miR-1-3p
Western blot was applied to explore the effect of LINC01518 knockdown or combined with miR-1-3p inhibition on the PIK3CA/Akt pathway in ESCC cells. The results implicated that LINC01518 silencing led to an evident reduction of PIK3CA and p-Akt expression in Ec9706 () and TE-1 () cells, while these effects were significantly overturned by anti-miR-1-3p. These data demonstrated that LINC01518 knockdown inhibited PIK3CA/Akt pathway in ESCC cells by regulating miR-1-3p.
Figure 6. Effects of LINC01518 knockdown or along with anti-miR-1-3p on the PIK3CA/Akt pathway in ESCC cells. (A and B) Ec9706 and TE-1 cells were cotransfected with si-LINC01518 or si-ctrl and anti-miR-1-3p or anti-miR-ctrl for 48 h, followed by Western blot analysis of PIK3CA, p-Akt and Akt protein levels. *p < .05.
Discussion
ESCC is believed to be one of the leading aggressive malignancies worldwide with high relapse and invasion rates, but the aetiology is still poorly understood. [Citation10]. Thus, a clear understanding of molecular mechanisms underlying the progression of ESCC is desperately needed for the improvement of ESCC survival rate. In recent years, an increasing number of studies have shown that dysregulated lncRNAs are widely implicated in the initiation and progression of ESCC [Citation11]. A variety of lncRNAs that are associated with ESCC initiation and progression have been characterized, such as LINC01419 [Citation12], small nucleolar RNA host gene 6 (SNHG6) [Citation13], antisense noncoding RNA in the INK4 locus (ANRIL) [Citation14], and growth arrest specific 5 (GAS5) [Citation15], and thus there has been an ongoing interest in the role of lncRNAs in ESCC [Citation16]. In our study, we presented another novel lncRNA, LINC01518, which was demonstrated to be highly expressed in ESCC tissues [Citation9]. We focussed on the roles of LINC01518 in ESCC progression. To the best of our knowledge, our study demonstrated for the first time that LINC01518 knockdown significantly suppressed cell proliferation and induced apoptosis in ESCC cells. Altogether, these results suggested that LINC01518 played an oncogenic role in ESCC progression.
miRNAs are a conserved group of endogenous small ncRNAs (approximately 19–24 nucleotides long) that negatively regulate their target gene expression at the post-transcriptional level [Citation17]. Over the past decades, the functions of miRNAs have been extensively studied in a variety of human malignancies including ESCC [Citation18,Citation19]. Therefore, numerous miRNAs have been identified as effective tumour biomarkers for cancer diagnosis in ESCC [Citation20]. Recently, a lncRNA-miRNA-mRNA regulatory circuitry has been proposed that lncRNAs may act as competing endogenous RNAs (ceRNAs) and competitively inhibit expression of miRNAs, eventually leading to the derepression of miRNA targets [Citation21]. For example, nuclear paraspeckle assembly transcript 1 (NEAT1) acted as an endogenous sponge to downregulate miR-129 by competitively binding to miR-129, thereby leading to the depression of its target gene C-terminal binding protein 2 (CTBP2) in ESCC cells [Citation22]. SNHG7 was documented to regulate the expression of inhibitor of DNA binding 4 (ID4) via sponging miR-342-3p in pancreatic cancer [Citation23]. To investigate whether LINC01518 was involved in the ceRNA regulatory network, we predicted the potential miRNAs that could be regulated by LINC01518. Bioinformatics analyses showed that miR-1-3p was one of the potential targets of LINC01518. Then, luciferase reporter assay and qRT-PCR proved that LINC01518 suppressed miR-1-3p expression by serving as a ceRNA for miR-1-3p. miR-1-3p, a muscle-specific miRNA, was reported to be frequently downregulated and identified as a tumour suppressive miRNA in many human tumours, such as bladder cancer [Citation24], prostate cancer [Citation24], hepatocellular carcinoma [Citation25], as well as ESCC [Citation26]. Our rescue experiments further manifested that miR-1-3p inhibition blocked the effects of LINC01518 knockdown on cell proliferation and apoptosis in ESCC cells. Collectively, the above results suggested that LINC01518 knockdown inhibited cell proliferation and induced apoptosis in ESCC cells by sponging miR-1-3p.
The PI3K/Akt pathway is one of the most crucial intracellular pathways which are essential for key biological processes such as cell growth, differentiation, apoptosis and metabolism, and reportedly contributes to tumour progression [Citation27]. Abnormal activation of the PI3K/Akt pathway, a common event in cancer progression, is mainly attributed to the accumulation of oncogenic activating mutations and tumour suppressor inactivation [Citation28]. PIK3CA, an extensively studied oncogene encoding the p110α catalytic subunit of PI3K, is frequently mutated and overexpressed in many different human malignancies including ESCC [Citation29,Citation30]. PIK3CA was demonstrated to be overexpressed in non-small-cell lung cancer (NSCLC) tissues and miR-1 targeted PIK3CA to inhibit tumorigenic properties in NSCLC cells [Citation31,Citation32]. More importantly, a recent study reported that miR-1 expression was lower in ESCC tissues and overexpression of miR-1 inhibited tumorigenicity of ESCC and enhanced the sensitivity of ESCC cells to anticancer drugs by suppressing PIK3CA/Akt pathway [Citation33]. Accordingly, our study supposed whether LINC01518 could regulate ESCC development by sponging miR-1-3p via inhibiting the PIK3CA/Akt pathway. In our study, we identified PIK3CA as a target of miR-1-3p in ESCC cells. Moreover, LINC01518 silencing significantly inhibited the PIK3CA/Akt pathway in ESCC cells, which was abolished by reintroduction with anti-miR-1-3p, indicating that LINC01518 knockdown inhibited the PIK3CA/Akt pathway by upregulating miR-1-3p. Collectively, we concluded that LINC01518 knockdown inhibited cell proliferation and induced apoptosis in ESCC cells via inhibiting the PIK3CA/Akt pathway by upregulating miR-1-3p.
Conclusion
In all, we provided the first evidence that LINC01518 expression was upregulated in ESCC cells and LINC01518 knockdown inhibited cell proliferation and induced apoptosis in ESCC cells by inhibiting the PIK3CA/Akt pathway via upregulating miR-1-3p. These findings suggested that targeting LINC01518/miR-1-3p/PIK3CA/Akt pathway might be an efficient therapeutic strategy for ESCC.
Disclosure statement
The authors declare that they have no conflict of interest.
References
- Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108.
- Morita FH, Bernardo WM, Ide E, et al. Narrow band imaging versus lugol chromoendoscopy to diagnose squamous cell carcinoma of the esophagus: a systematic review and meta-analysis. BMC Cancer. 2017;17(1):54.
- Hiyama T, Yoshihara M, Tanaka S, et al. Genetic polymorphisms and esophageal cancer risk. Int J Cancer. 2007;121(8):1643–1658.
- Markar SR, Wiggins T, Ni M, et al. Assessment of the quality of surgery within randomised controlled trials for the treatment of gastro-oesophageal cancer: a systematic review. Lancet Oncol. 2015;16(1):e23–31.
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30.
- Bertone P, Stolc V, Royce TE, et al. Global identification of human transcribed sequences with genome tiling arrays. Science. 2004;306(5705):2242–2246.
- Deng G, Sui G. Noncoding RNA in oncogenesis: a new era of identifying key players. IJMS. 2013;14(9):18319–18349.
- Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81(1):145–166.
- Wang W, Wei C, Li P, et al. Integrative analysis of mRNA and lncRNA profiles identified pathogenetic lncRNAs in esophageal squamous cell carcinoma. Gene. 2018;661:169–175.
- Liu SY, Chen W, Chughtai EA, et al. PIK3CA gene mutations in Northwest Chinese esophageal squamous cell carcinoma. WJG. 2017;23(14):2585–2591.
- Sugihara H, Ishimoto T, Miyake K, et al. Noncoding RNA expression aberration is associated with cancer progression and is a potential biomarker in esophageal squamous cell carcinoma. IJMS. 2015;16(11):27824–27834.
- Chen JL, Lin ZX, Qin YS, et al. Overexpression of long noncoding RNA LINC01419 in esophageal squamous cell carcinoma and its relation to the sensitivity to 5-fluorouracil by mediating GSTP1 methylation. Ther Adv Med Oncol. 2019;11:175883591983895.
- Zhang Y, Li R, Ding X, et al. Upregulation of long non-coding RNA SNHG6 promote esophageal squamous cell carcinoma cell malignancy and its diagnostic value. Am J Transl Res. 2019;11(2):1084–1091.
- Cao T, Shen J, Pan W, et al. Upregulation of long noncoding RNA ANRIL correlates with tumor progression and poor prognosis in esophageal squamous cell carcinoma. J Buon. 2018;23(6):1862–1866.
- Wang G, Sun J, Zhao H, et al. Long non-coding RNA (lncRNA) growth arrest specific 5 (GAS5) suppresses esophageal squamous cell carcinoma cell proliferation and migration by inactivating phosphatidylinositol 3-kinase (PI3K)/AKT/mammalian target of rapamycin (mTOR) signaling pathway. Med Sci Monit. 2018;24:7689–7696.
- Yao J, Huang JX, Lin M, et al. Microarray expression profile analysis of aberrant long non-coding RNAs in esophageal squamous cell carcinoma. Int J Oncol. 2016;48(6):2543–2557.
- Schickel R, Boyerinas B, Park SM, et al. MicroRNAs: key players in the immune system, differentiation, tumorigenesis and cell death. Oncogene. 2008;27(45):5959–5974.
- Martens-Uzunova ES, Bottcher R, Croce CM, et al. Long noncoding RNA in prostate, bladder, and kidney cancer. Eur Urol. 2014;65(6):1140–1151.
- David S, Meltzer SJ. MicroRNA involvement in esophageal carcinogenesis. Curr Opin Pharmacol. 2011;11(6):612–616.
- Kimura S, Naganuma S, Susuki D, et al. Expression of microRNAs in squamous cell carcinoma of human head and neck and the esophagus: miR-205 and miR-21 are specific markers for HNSCC and ESCC. Oncol Rep. 2010;23(6):1625–1633.
- Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154(1):26–46.
- Li Y, Chen D, Gao X, et al. LncRNA NEAT1 regulates cell viability and invasion in esophageal squamous cell carcinoma through the miR-129/CTBP2 axis. Dis Markers. 2017;2017:1.
- Cheng D, Fan J, Ma Y, et al. LncRNA SNHG7 promotes pancreatic cancer proliferation through ID4 by sponging miR-342-3p. Cell Biosci. 2019;9(1):28.
- Wang W, Shen F, Wang C, et al. MiR-1-3p inhibits the proliferation and invasion of bladder cancer cells by suppressing CCL2 expression. Tumour Biol. 2017;39:1010428317698383.
- Zhang H, Zhang Z, Gao L, et al. miR-1-3p suppresses proliferation of hepatocellular carcinoma through targeting SOX9. OTT. 2019;12:2149–2157.
- Liu W, Li M, Chen X, et al. MicroRNA-1 suppresses proliferation, migration and invasion by targeting Notch2 in esophageal squamous cell carcinoma. Sci Rep. 2018;8(1):5183.
- Hennessy BT, Smith DL, Ram PT, et al. Exploiting the PI3K/AKT pathway for cancer drug discovery. Nat Rev Drug Discov. 2005;4(12):988–1004.
- Courtney KD, Corcoran RB, Engelman JA. The PI3K pathway as drug target in human cancer. JCO. 2010;28(6):1075–1083.
- Jung K, Kang H, Mehra R. Targeting phosphoinositide 3-kinase (PI3K) in head and neck squamous cell carcinoma (HNSCC). Cancers Head Neck. 2018;3(1):3.
- Akagi I, Miyashita M, Makino H, et al. Overexpression of PIK3CA is associated with lymph node metastasis in esophageal squamous cell carcinoma. Int J Oncol. 2009;34:767–775.
- Yu QQ, Wu H, Huang X, et al. MiR-1 targets PIK3CA and inhibits tumorigenic properties of A549 cells. Biomed Pharmacother. 2014;68(2):155–161.
- Zhao Q, Zhang B, Shao Y, et al. Correlation between the expression levels of miR-1 and PIK3CA in non-small-cell lung cancer and their relationship with clinical characteristics and prognosis. Future Oncol. 2014;10(1):49–57.
- Yu Q, Liu Y, Wen C, et al. MicroRNA-1 inhibits tumorigenicity of esophageal squamous cell carcinoma and enhances sensitivity to gefitinib. Oncol Lett. 2018;15:963–971.
