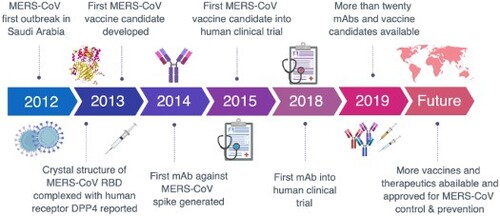
ABSTRACT
The Middle East respiratory syndrome coronavirus (MERS-CoV) has spread through 27 countries and infected more than 2,200 people since its first outbreak in Saudi Arabia in 2012. The high fatality rate (35.4%) of this novel coronavirus and its persistent wide spread infectiousness in animal reservoirs have generated tremendous global public health concern. However, no licensed therapeutic agents or vaccines against MERS-CoV are currently available and only a limited few have entered clinical trials. Among all the potential targets of MERS-CoV, the spike glycoprotein (S) has been the most well-studied due to its critical role in mediating viral entry and in inducing a protective antibody response in infected individuals. The most notable studies include the recent discoveries of monoclonal antibodies and development of candidate vaccines against the S glycoprotein. Structural characterization of MERS-CoV S protein bound with these monoclonal antibodies has provided insights into the mechanisms of humoral immune responses against MERS-CoV infection. The current review aims to highlight these developments and discuss possible hurdles and strategies to translate these discoveries into ultimate medical interventions against MERS-CoV infection.
1. Introduction
The rapid emergence and dissemination of infectious diseases has taken a heavy toll on humans since the beginning of the twenty-first century. One of the most well-known examples was the outbreak of severe acute respiratory syndrome (SARS) in the winter of 2002 and 2003, caused by a novel coronavirus (SARS-CoV) [Citation1,Citation2]. In distinct contrast to the mild human coronaviruses HCoV-229E [Citation3], HCoV-OC43 [Citation4], HCoV-NL63 [Citation5], and HCoV-HKU1 [Citation6], infection with SARS-CoV frequently resulted in severe symptoms including fever, dry cough, shortness of breath and pneumonia. Transmission of SARS-CoV was primarily from person to person and most cases occurred in health care settings lacking adequate infection control precautions [Citation2]. The SARS outbreak had severe consequences in 29 countries and regions, infecting 8096 people worldwide with a fatality rate of approximately 10% [Citation7]. There are still no vaccines or therapeutics specific to SARS-CoV available 16 years after the SARS outbreak. It is not hard to imagine how catastrophic it would be if SARS-CoV were to hit the human community again.
While SARS-CoV remains a mystery and a loose cannon, another novel coronavirus emerged in Saudi Arabia in 2012, later known as the Middle East respiratory syndrome coronavirus (MERS-CoV) [Citation8]. The fatality rate of MERS-CoV infection is approximately 35.4%, and new cases as well as associated deaths continue to arise to date [Citation9]. Despite that most cases have been attributed to human-to-human transmission, MERS-CoV does not appear to transmit efficiently among humans unless there is close contact. The exact source of MERS-CoV and its routes of transmission to humans still remain uncertain. Dromedary camels are believed to be the animal reservoir for MERS-CoV because isolates from camels are almost identical to those from human, and that many domestic camels are seropositive for MERS-CoV (reviewed in [Citation10,Citation11]). Furthermore, current evidence strongly suggests that bats are the original source for MERS-CoV, as many coronaviruses phylogenetically related to MERS-CoV originate in bats, including BatCoV-HKU4, BatCoV-HKU5 and other MERS-related coronaviruses [Citation12–15]. The BatCoV-HKU4 was also shown to be able to engage the cellular receptor of MERS-CoV, adding evidence to the bat origin theory [Citation16]. However, there has not yet been direct evidence for isolating MERS-CoV from bats (reviewed in [Citation10,Citation11,Citation17]).
Great efforts have been made to develop preventive and therapeutic interventions against MERS-CoV infection. In particular, monoclonal antibodies and vaccines targeting the Spike glycoprotein are major areas of focus due to its critical role in mediating viral entry, and its potential in inducing protective antibody responses in infected individuals. So far, more than twenty monoclonal antibodies with nanomolar neutralizing activities have been reported and many vaccine candidates are underway in preclinical and clinical studies. In this review, we aim to capture the current advances and discuss possible strategies to translate these discoveries into an ultimate medical intervention against MERS-CoV infection.
2. Structure and function of MERS-CoV spike glycoprotein
MERS-CoV belongs to the genus betacoronavirus of the coronaviridae family [Citation18]. It is an enveloped, single-stranded, positive-sense RNA virus with a helical capsid structure ((A)). The genome of MERS-CoV is around 30 kb (30,119nt) long and encodes 4 structural proteins (Spike, Envelope, Membrane, and Nucleocapsid) and 16 nonstructural proteins ((C)) [Citation13]. Like other coronaviruses, the MERS-CoV uses its spike (S) glycoprotein to interact with cellular receptors and enter into the target cell [Citation19–22]. As a unique structural component of the virion membrane, the S glycoprotein assembles into trimers and forms large protruding spikes on the surface of the virion [Citation20]. The S glycoprotein is a typical type I membrane glycoprotein consisting of a globular S1 domain at the N-terminal, followed by a membrane-proximal S2 domain and a transmembrane (TM) domain [Citation21]. The S1 domain mediates viral attachment and contains the RBD (receptor binding domain), which determines the host range and cellular tropism for MERS-CoV [Citation23–25]. Similar to other coronaviruses, the S2 domain of MERS-CoV, mediating membrane fusion, contains the hydrophobic fusion peptide (FP) at the N-terminus as well as two heptad repeats designated as HR1 and HR2 ((C)) [Citation26]. Through co-purification with the MERS-CoV S1 domain, Raj and colleagues identified that dipeptidyl peptidase 4 (DPP4, also known as CD26) functions as a cellular receptor for MERS-CoV [Citation27].
Figure 1. General introduction to MERS-CoV: model structure, life cycle and genomic composition. (A) Cartoon model structure of MERS-CoV. (B) Membrane fusion mechanism for MERS-CoV spike glycoprotein. Binding between RBD and the cell receptor (DPP4) triggers the conformational change of S glycoprotein to form a pre-hairpin intermediate of S2, in which the hydrophobic HR1 is exposed and the fusion peptide inserts into the target cell membrane. This transient S2 intermediate then refolds with HR2 into a stabilized trimer of hairpins, also called six-helix bundle structure (6-HB), bringing the target cell membrane into close proximity of the viral envelope and resulting in the completion of the fusion process. (C) Genomic composition of MERS-CoV. Each coloured box (length in scale) represents one open reading frame in the genomic RNA. The schematic for spike glycoprotein was also shown with labelled domain names and residue numbers. ORF (open reading frame), DPP4 (dipeptidyl peptidase 4), RBD (receptor-binding domain), NTD (N-terminal domain), CTD (C-terminal domain), FP (fusion peptide), and HR1-2 (heptad repeats 1-2).
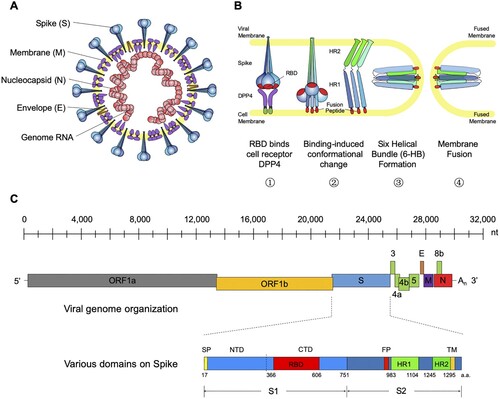
The MERS-CoV virion enters the host airway cells in the respiratory tract through fusion with either the plasma or endosomal membrane [Citation19]. Binding between RBD and the cell receptor triggers a cascade of conformational changes that lead to the formation of a pre-hairpin intermediate of S2, in which the hydrophobic HR1 is exposed and allows the fusion peptide to insert into the target cell membrane. This transient S2 intermediate then refolds with HR2 into a stabilized trimer of hairpins, also called six-helix bundle structure (6-HB), which brings the target cell membrane into close proximity of the viral envelope, resulting in the completion of the fusion process and initiation of the virus life cycle [Citation21] ((B)). Structure-based design of various peptides able to block the formation of 6-HB have demonstrated potent inhibition on MERS-CoV replication and spike-mediated cell–cell fusion, showing great promise for further development into effective viral fusion inhibitors for treating MERS-CoV infection [Citation26,Citation28–30]. Among them, the peptide EK1 is effective to multiple human coronaviruses apart from MERS-CoV and therefore serves as a potential pan-coronavirus fusion inhibitor [Citation30].
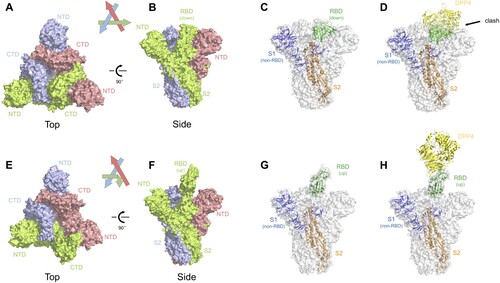
Recently, structural studies on the prefusion state spike protein of MERS-CoV and SARS-CoV have provided more insights into the spike-mediated membrane fusion process [Citation31–34]. The MERS-CoV spike protein trimerizes and folds into a metastable prefusion conformation on the virion surface, in which three S1 domains fold into a steady trimer structure and sit on top to stabilize the coiled S2 domains ((A–B)). We and others have identified that the RBD of SARS-CoV and MERS-CoV can be found either buried (‘down' position) or exposed (‘up' position) in the spike trimer structure [Citation31,Citation33–35]. The two conformational states of RBD may have distinct roles during receptor binding and membrane fusion: only the RBDs in ‘up' position, but not those in ‘down' position, can bind to the cell receptor DPP4 ((C–D)). Great steric clash was observed between DPP4 and neighboring spike protomers when we mapped it to the RBD in ‘down' position ((C–D)). Transformation of the RBD from the buried to the exposed state is therefore a prerequisite for receptor binding ((G–H)). On the other hand, this conformational change also seems to open up the stable cap structure sitting above the S2 cores ((E–F)). This may lead to disassociation of S1 trimer and exposure of the fusion apparatus, triggering the membrane fusion process.
Figure 2. Structural insights of the MERS-CoV spike glycoprotein. (A–B) Top and side view of the MERS-CoV spike trimer with all RBD in ‘down' position, shown as molecular surface (PDB ID: 5W9J). The three protomers are coloured green, lightblue, and red, respectively. The labels are the same with those in . (C) One of the three protomers in (B) is highlighted as cartoon representation whereas the other two protomers are faded in white. The RBD in ‘down' position is coloured in green. The non-RBD S1 region was coloured deep blue and the S2 region was coloured orange. (D) Superimposition of RBD-bound DPP4 (PDB ID: 4L72) into the MERS-CoV spike trimer. Clashes were observed between DPP4 and the other two S1 regions in the trimer structure. (E–F) Top and side view of the MERS-CoV spike trimer with one RBD in ‘up' position and the other two in ‘down' position, shown as molecular surface (PDB ID: 5W9H). Same colour codes are used as in (A–B). (G) The protomer with RBD in ‘up' position is highlighted as cartoon representation, with RBD in green, non-RBD S1 region in deep blue, and S2 in orange. (H) Superimposition of the RBD-bound DPP4 into the MERS-CoV spike trimer, with DPP4 interacting with the ‘up' RBD. No steric clash was observed.
To gain a better understanding of MERS-CoV interaction with cellular receptors at atomic levels, we and others have determined the crystal structure of MERS-CoV RBD bound to the extracellular domain of its cellular receptor dipeptidyl peptidase 4 (DPP4) [Citation23,Citation24]. We showed that MERS-CoV RBD consists of a core and a receptor binding subdomain. MERS-CoV RBD and the related SARS-CoV RBD share a high degree of structural similarity in their core subdomains, but are notably divergent in the receptor binding subdomains [Citation36]. The receptor binding subdomain of MERS-CoV RBD directly interacts with blades 4 and 5 of DPP4 propeller instead of its intrinsic hydrolase domain. The interface consists of a buried surface of ∼2550 Å2 involving 14 residues in receptor binding subdomain interacting with 15 residues in DPP4. The actual binding forces are mediated through two major binding patches. Patch 1 represents 49% of buried surface and forms between the C-terminal end of the long loop connecting the β6 and β7 strands and blade 4 of DPP4. Patch 2 occupies 51% of buried surface and forms a slightly concaved outer surface at the far end of the MERS-CoV receptor binding subdomain and a linker containing a short helix between blade 4 and blade 5 of DPP4. The concaved outer surface is made by the short β6 strand, C-terminal parts of β5 and β7 strands, N-terminal part of β8 strand and the β5-β6 linking loop. It is hoped that better understanding of the atomic details of the spike glycoprotein, as well as the interface between MERS-CoV RBD and DPP4 will provide the structural basis for rational design and development of therapeutics and vaccines against MERS-CoV infection.
3. Neutralizing monoclonal antibodies against MERS-CoV infection
Neutralizing antibodies are a major component of protective immunity against viral infection in humans. Polyclonal by nature, the antibody response in vivo mobilizes a dynamic and complex mixture of monoclonal antibodies (mAbs) that work in concert to target various antigenic domains on the viral envelope glycoprotein. Identifying the neutralizing mAbs that constitute the neutralizing activity of polyclonal response and their recognized antigenic domains has therefore become the first crucial step towards gaining a better understanding of the protective antibody response, developing clinical intervention methods, and designing immunogens capable of eliciting neutralizing antibodies.
Great achievements have been made in the isolation of neutralizing mAbs in the past few years using various technology platforms (). Up till now, more than 20 mAbs, most of which are human or humanized antibodies, have been described by scientists from all over the world. These antibodies are listed in chronological order of publication in , together with their unique biochemical and antiviral properties against MERS-CoV infection observed in cell culture and experimental animal models.
Figure 3. Development of monoclonal antibodies against MERS-CoV. (A) Monoclonal antibodies sorted from non-immunized human scFv (single-chain fragment variable) libraries. MERS-4 and MERS-27 were isolated from a non-immunized human scFv library displayed on yeast with MERS-CoV spike RBD as bait protein. Similarly, 3B11 and m336 were isolated from non-immunized human scFv phage libraries with MERS-CoV S protein or RBD protein as bait protein, respectively. (B) Monoclonal antibodies sorted from immunized animals. The antibodies 5F9, hMS-1 (Mersmab-1), D12, F11, G2, G4, 4C2h (4C2), REGN3048 and REGN3051 were isolated from mice immunized with the indicated vaccines labelled in the colour-coded boxes, each representing a different immunogen; the bait or target protein for antibody selection were also listed. The mice from which REGN3048 and REGN3051 were isolated were given the pale blue colour to indicate that they express human immunoglobulin genes. NbMS10-Fc, JC57-11, JC57-13, JC57-14, F1B-H1 and HCAb-83 were isolated from larger animal including llama, rhesus macaque and camels as indicated. The vaccines and selection criteria were also shown. NTD (N-terminal domain), Fc (fragment constant). (C) Monoclonal antibodies isolated from human survivors recovered from MERS-CoV infection. MERS-GD27, MERS-GD33, LCA60, CDC-C2, CDC-C5, CDC-A2 and CDC-A10 were generated by culturing B cells sorted from the patient and screening for MERS-CoV-specific antibodies. MCA1 was produced by constructing a phage library displaying scFv cloned from a convalescent patient.
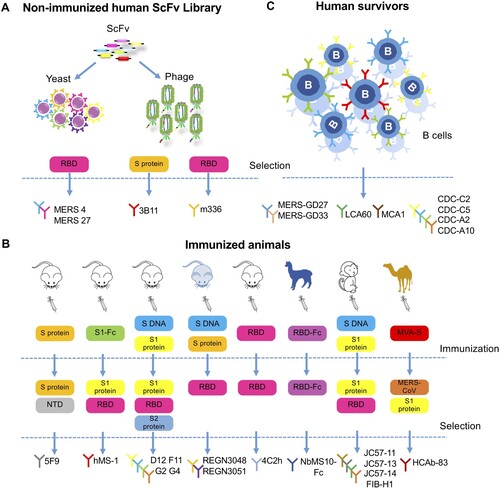
Table 1. Advancement in MERS-CoV monoclonal antibodies development.
It is apparent that the single chain fragment variable (scFv) library approach allows rapid discovery of mAb, without time constraints from immunizing experimental animals or approaching convalescent individuals of MERS-CoV infection. The earliest mAbs reported in 2014 were identified through screening non-immune human scFv libraries with the ectodomain of S glycoprotein (mAb 3B11) [Citation40] or soluble RBD from S glycoprotein (MERS-4, MERS-27 and the m336 panel) [Citation37,Citation42] as bait protein ((A)). These antibodies all demonstrated high neutralizing activities and therefore were widely used as reference antibodies in later studies.
Antibodies have also been generated from immunized animals ((B)). Several groups have reported mAbs isolated from either wild-type inbred mice or transgenic mice expressing human antibody-variable heavy chains and κ light chains. Mersmab-1 (known as hMS-1 after humanization) was isolated from mice immunized subcutaneously with chimeric S1-Fc [Citation47,Citation48]. The mAbs 2E6 and 4C2 (humanized form 4C2 h) were isolated in mice immunized with recombinant RBD produced in insect cells [Citation55]. Furthermore, two human-like mAbs, REGN3048 and REGN3051, were directly cloned from transgenic mice expressing human versions of the antibody after immunization with DNA encoding S glycoprotein and purified recombinant S glycoprotein [Citation51]. Both mAbs have been tested in humanized mice models and in non-human primates [Citation51,Citation52]. The authors indicated that the advantages of their system not only lay in the human component of their antibodies but also in the quick speed associated with isolation and production, since no humanization or optimization step was required. Currently, REGN3048 and REGN3051 have entered phase I clinical trials.
Most of the mAbs reported so far target the RBD region of S glycoprotein, but RBD does not seem to be the only target for anti-MERS-CoV antibody responses. Recently, a mAb targeting the S1 N-terminus domain (NTD) region, which does not contain RBD, was isolated from mice immunized with S glycoprotein [Citation57]. This antibody, 5F9, was shown to successfully block virus entry in cell culture models and the efficacy was comparable to other mAbs in IC50. Further, the mAb panel D12, F11, G2 and G4 were generated by priming mice with DNA encoding the full-length S glycoprotein and boosting them with S1 protein. Among them are two mAbs that target the non-RBD S1 (mAb G2) and S2 region (mAb G4), respectively [Citation49]. These non-RBD-binding antibodies potently neutralized pseudo- and live MERS-CoV in cell culture and were also protective in mouse models [Citation49,Citation50]. Together, the development of these antibodies elucidates that RBD may not be the single target for anti-viral antibody response. More studies are needed to elaborate the detailed mechanisms for these antibodies.
Apart from the traditional approach of isolating mAbs from immunized mice, several groups have turned to larger animal models for antibody isolation. One group immunized rhesus macaques with combined DNA and protein vaccines and isolated a panel of mAbs, including JC57-11, JC57-13, JC57-14, and FIB-H1, targeting both RBD and non-RBD S1 region of the S glycoprotein, all with potent neutralizing activities [Citation50]. Another group immunized llama with recombinant RBD and screened the nanobody library for high-affinity single heavy chain antibody (nanobody) against RBD. The humanized form NbMS10-Fc was constructed by combining the variable domain of the nanobody with the human constant Fc domain, and it was shown to protect mice from lethal MERS-CoV challenge [Citation60]. Similarly, Stalin et al isolated a nanobody targeting RBD from camels immunized with MVA encoding S glycoprotein. The humanized form HCAb-83 has high binding affinity to S protein and potent neutralizing activities to live virus [Citation61]. These nanobody-derived mAbs are smaller in molecular weight and more stable than traditional antibodies, and may provide a new option for future antibody isolation.
In terms of closeness to authentic human antibodies, no approach can compete with those based on direct B cell cloning from convalescent individuals. One such mAb LCA60 was isolated from memory B cells of human survivors of MERS-CoV infection and was among the most potent mAbs reported in neutralizing pseudo- and live viruses [Citation53]. More mAbs isolated from human survivors were described as more convalescent blood samples became available, including MCA1 [Citation56], CDC-C2, CDC-C5, CDC-A2, CDC-A10 [Citation50], MERS-GD27, and MERS-GD33 [Citation58,Citation59] ((C)), all with potent neutralizing activities against MERS-CoV. The mAbs LCA60, CDC-C2, MCA1, and MERS-GD27 were also tested to be protective in animal models.
As MERS-CoV research progressed quickly in the past few years, many mAbs have been tested for prophylactic or therapeutic protection efficacy in human DPP4 transgenic / transduced mice models, and a few have entered large animal model trials such as in rabbits or non-human primates (NHPs). However, as different animal models were established among labs worldwide with slightly different evaluation end points, it is difficult to make a direct comparison among these mAbs. This is also true for in vitro evaluation of neutralizing activities – since different cell lines, pseudo-viruses, and neutralizing assay techniques are utilized, the published IC50 values can only serve as indirect reference for comparison. Head to head comparison in the same experimental system would be required to identify the most protective mAb or combination of mAbs against MERS-CoV infection in order to proceed to clinical trials.
4. Structure features of neutralizing mAbs against MERS-CoV infection
We and others have carried out structural studies of MERS-CoV neutralizing antibodies in complex with MERS-RBD to understand neutralizing mechanism at atomic levels (). Based on the epitopes revealed by structural studies, MERS-CoV antibodies targeting RBD can be classified into three groups ((B), ).
Figure 4. Advancement in structural studies of MERS-CoV neutralizing monoclonal antibodies. (A) Structure of MERS-CoV spike trimer ectodomain (PDB ID: 5X5F). A single protomer of the trimeric spike protein with RBD in ‘up' conformation is shown as molecular surface. The RBD, NTD and S2 subunit are coloured in green, paleyellow and lightblue, respectively. The two remaining protomers with RBD in ‘down' conformation are shown in cartoon representation and coloured in wheat. (B) Structures of MERS-CoV neutralizing antibodies targeting RBD. Antibodies are classified into three groups and shown as cartoon representation. The RBD is coloured in green and antibodies in different colours. Group 1 includes MERS-27 (PDB ID: 4ZS6), D12 (PDB ID: 4ZPT), 4C2 (PDB ID: 5DO2) and JC57-14 (PDB ID: 6C6Y). Group 2 includes m336 (PDB ID: 4XAK), MCA1 (PDB ID: 5GMQ), CDC2-C2 (PDB ID: 6C6Z) and MERS-GD27. Group 3 includes MERS-4 (PDB ID: 5ZXV) and MERS-4V2 (PDB ID: 5YY5). (C) Neutralizing mechanisms of MERS-CoV neutralizing antibodies targeting RBD. The left panel shows the structural superimposition of the representative antibodies from the three groups (MERS-27, m336 and MERS-4) and DPP4 (coloured in yellow) bound to RBD (coloured in green, PDB ID: 4L72) at the same time. The right panel is enlarged view of steric clashes between the antibodies and the DPP4 and a significant conformational difference in the RBD β5-β6 loop between antibody-bound and DPP4-bound states. (D) Structure of G4 fab (coloured in teal) in complex with spike trimer (PDB ID: 5W9H). (E) Side views of spike monomer bound to G4 fab. The enlarged view of the glycosylated loop in the S2 subunit recognized by G4 is shown on the right.
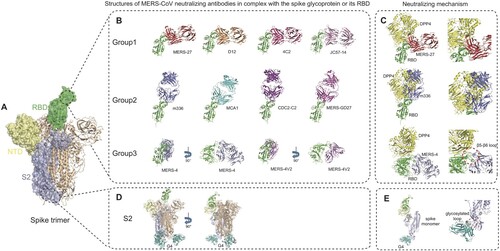
The first group consists of antibodies MERS-27, D12, 4C2 and JC57-14, which interact with the C-terminal segment of the β6-β7 loop and β7 strand of RBD by both heavy and light chains ((B)) [Citation38,Citation49,Citation50,Citation55]. Their common epitopes on the RBD include residues Val527, Ser528, Ile529, Val530, Pro531, Ser532, Trp535, Glu536 and Asp539 in the β6-β7 loop. The residues Trp535, Glu536 and Asp539 also happen to be within the DPP4-binding site patch 1 of MERS-CoV RBD [Citation23], mediating interaction with Lys267 and the carbohydrate moiety linked to Asn229 of DPP4 [Citation38]. Therefore, the Group 1 antibodies would directly compete with DPP4 in binding to RBD by interfering with both protein–protein and protein–carbohydrate interactions between RBD and DPP4. Structural super-impositions also showed that these four antibodies and DPP4 would have steric clashes between the variable domain of the heavy chain and the propeller domain of DPP4 if they simultaneously bind to RBD ((C)).
The second group consists of antibodies m336, MCA1, CDC2-C2 and MERS-GD27, which interact with the β5-β8 strands, β5-β6 loop and β6-β7 loop in RBD mainly by the heavy chain ((B)) [Citation43,Citation50,Citation56,Citation58]. Their common epitope consists of Phe/Leu506, Asp510, Trp535, Glu536, Asp539, Tyr540, Tyr541, Arg542, and Trp553. Although antibodies in both Group 1 and Group 2 share the binding residues Trp535, Glu536 and Asp539, their approaching angles to the RBD are significantly different. As shown in (C), the approaching angle of Group 2 antibodies is closer to that of DPP4 by rotating approximately 90 degrees anti-clockwise from that of Group 1 antibodies, thereby generating more steric clashes with DPP4. This is also evidenced by a larger overlap between the common epitope of Group 2 antibodies and DPP4-binding site on RBD [Citation23]. As a representative of Group 2 antibodies, m336 exhibits very potent neutralizing activity by not only mimicking critical interactions between RBD and DPP4 but also adopting an approaching angle similar to that of DPP4 ((C)).
The third group consists of antibody MERS-4 and its variant MERS-4V2 with four residue replacements in the HCDR3 ((B)) [Citation39]. By structural determination, it was shown that MERS-4 Fab and MERS-4V2 scFv share the same mode of binding to the RBD ((B)) [Citation39]. Analysis of the RBD/MERS-4V2 complex structure showed that the antibody contacts with the β5-β6, β6-β7 and β7-β8 loops of the receptor-binding subdomain in RBD [Citation39]. The epitope involves Leu507, Ser508, Gln516, Asn519, Asn521, Gln522, Tyr523, Pro525, Lys543, Leu545, and Gly550 [Citation39]. To be note, the MERS-4 epitope has no overlap with DPP4-binding site ((C)). By approaching the RBD outside the DPP4-binding site, MERS-4 recognizes a unique epitope different from all previously reported RBD-targeting antibodies. Comparisons of RBD in DPP4-bound and MERS-4-bound states revealed that binding of MERS-4 induces or fixes the β5-β6 loop into a conformation in which it folds into a shallow groove on the RBD interface critical for accommodating a short helix of DPP4, thereby indirectly disrupting the interaction between RBD and DPP4 ((C)). Such different epitope and mechanism enable MERS-4 to synergize with other antibodies including RBD-targeting MERS-27 and m336 in neutralization, which provides valuable addition for the combined use of antibodies against MERS-CoV infection [Citation39].
In addition to the aforementioned ten antibodies targeting RBD, the near atomic resolution cryo-EM structures of the trimeric MERS-CoV spike and its complex with antibody G4 were also determined ((D)) [Citation33]. G4 is the first reported S2-targeting antibody and its epitope consists of a glycosylated, solvent-exposed loop residing in a connector domain between the HR1 and HR2 of the S2 subunit. In the unbound spike trimer structure this loop is largely disordered, whereas it extends out from two β-strands and is surrounded by all six CDRs (complementarity determining regions) of the mAb G4 upon antibody recognition ((E)). The specific spike-G4 interaction may stabilize the loop and further impede conformational changes of S2 subunit essential for membrane fusion after DPP4 binding. The binding epitope for G4 in S2 subunit is more conserved than RBD among MERS-CoV isolates, shedding light on G4 as a potential broad-spectrum neutralizing antibody for MERS-CoV. Yet this loop between HR1 and HR2 is variable in sequence and length among different viruses even in lineage C betacoronaviruses [Citation33], limiting its application to other coronaviruses. In terms of pan-coronavirus medical countermeasures (MCMs), the recently developed fusion inhibitor peptide EK1 is a potential candidate. The peptide EK1 was designed to target the more conserved HR1 region of the S2 stem, and was shown to block cell–cell fusion induced by spike protein from multiple human coronaviruses [Citation30].
In general, most reported MERS-CoV neutralizing antibodies recognize the RBD in the S1 subunit, and these antibodies are highly potent in neutralization. These facts show that the RBD in the S1 subunit is a major vulnerable site for antibody recognition and neutralization. To be note, the RBD is also the region where most naturally occurring mutations of the S glycoprotein occur. Currently, the comprehensively studied antibodies targeting the non-RBD region of the spike glycoprotein also include mAb 5F9 targeting the N-terminal domain (NTD) of the S1 subunit [Citation57], as well as mAbs G2, CDC2-A2, CDC2-A10, JC57-13 and FIB-H1 targeting the non-RBD region of the S1 subunit [Citation33,Citation50]. However, the detailed epitopes and specific mechanisms are still unclear for these antibodies. We expect that more antibodies with new neutralizing epitopes and/or mechanisms would be important for the combined use of antibodies against MERS-CoV infection.
5. Advancement in MERS-CoV vaccine development
Although monoclonal antibodies show promising anti-viral effects in both cell culture and animal models against MERS-CoV infection, their roles are still limited in large-scale disease prevention in MERS-CoV high risk areas, as the therapeutic window is generally narrow for mAbs and mass-scale production is time- and resource-consuming. Vaccines still remain the best choice for MERS-CoV prevention.
Given its critical role in mediating viral entry and as major targets for neutralizing antibodies, S glycoprotein and its RBD have become the prime targets for MERS-CoV immunogen design and vaccine development. Various approaches have been applied and more than twenty vaccine candidates have been reported in the past few years, including vaccines based on inactivated virions [Citation62,Citation63], virus-like particles [Citation64], recombinant viral vectors [Citation65–80], DNA [Citation49,Citation81,Citation82], recombinant protein subunits [Citation33,Citation49,Citation83–92], and nanoparticles [Citation80,Citation93,Citation94]. summarizes the critical features of these approaches and their protective potentials in experimental animal models.
Table 2. Advancement in MERS-CoV vaccine development.
Up till now, only two vaccine candidates, GLS-5300 and MERS001, have entered human clinical trials. The vaccine GLS-5300 was the first to be tested in healthy human volunteers. It is a DNA plasmid encoding the MERS-CoV S glycoprotein, requiring two-to-three injections delivered by electroporation [Citation81]. The phase I clinical trial was started in 2016 at the Walter Reed Army Institute, and another phase I/II clinical trial is being conducted in Korea to test dosage safety and immunogenicity. Another vaccine candidate, MERS001, is a replication-deficient chimpanzee adenovirus (ChAdOx1) containing the MERS-CoV S glycoprotein antigen [Citation70,Citation71]. This vaccine only requires one-time administration of 5×109–5×1010 virus particles via intramuscular route, and the local adverse events as well as immunogenicity will be evaluated in the phase I clinical trial conducted at the University of Oxford. In addition, one more candidate vaccine has been tested in dromedary camels either for potential human use or straight into veterinary use. It explores a modified vaccinia virus Ankara (MVA) as a vector to express MERS-CoV S glycoprotein [Citation67]. The regimen involves immunization through intranasal as well as intramuscular routes twice at a 4-week interval. The vaccinated camels demonstrated a significant reduction of excreted infectious virus and viral RNA transcripts in vaccinated animals upon MERS-CoV challenge. Protection against MERS-CoV infection correlated with the presence of serum neutralizing antibodies to MERS-CoV. As MVA has established a reasonably good safety profile in humans and induced desirable protective immunity in camels, it represents one of the potential candidates to be further evaluated in humans in the near future.
The remaining vaccine candidates are all in the stages of preclinical or laboratory development and invariably target the S glycoprotein or RBD critical for viral entry (). Vaccines based on inactivated [Citation62,Citation63] or virus-like particles [Citation64] have historical precedence in inducing protective immune responses in humans. Whether the same strategies are applicable to MERS-CoV requires further studies, particularly when it comes to possible safety concerns [Citation62].
Apart from MERS001 and the MVA-based vaccine tested in dromedary camels, other vector-based approaches are also being actively pursued, including adenovirus [Citation68,Citation69,Citation72,Citation73,Citation80], measles virus [Citation74,Citation75], VEEV replicon particle [Citation76,Citation77], vesicular stomatitis virus [Citation78], and rabies virus [Citation79]. All recombinant viruses encoding the MERS-CoV S or S1 antigen demonstrated strong immunogenicity in mice or non-human primate models, and some were shown to confer protection in MERS-CoV challenge mouse models (). However, concerns remain regarding the pre-existing immunity against these viral vectors from natural infection, because it would diminish the vaccine potency [Citation95]. To overcome the issue of pre-existing immunity against human adenoviruses while preserving their advantages such as high yields and strong immunogenicity, rare serotypes of chimpanzee adenovirus of low human seroprevalence may be adopted as viral vectors [Citation70,Citation73]. Our group recently developed a vaccine candidate with replication-defective chimpanzee adenovirus C68 (AdC68) vector expressing full length MERS-CoV S glycoprotein. Seroprevalence of AdC68 is around 2% in human population, much lower than that of the commonly used human adenovirus 5 (HuAd5) vector (>60%) [Citation96,Citation97]. One intra-nasal administration of 2 × 109 viral particles completely protected human DPP4 knock-in (hDPP4-KI) mice from lethal MERS-CoV challenge, and passive transfer of AdC68-S immune sera conferred survival advantage in lethal challenge mouse models [Citation73]. Further, the safety profiles of these vectors have yet to be extensively tested in humans. Recently, Hashem and colleagues showed that the adenovirus-based S1 vaccine may pose potential safety concerns because it may induce pulmonary perivascular hemorrhage in a MERS-CoV challenge mouse model, regardless of the its full protection upon lethal viral infection. They also showed that the pulmonary pathology can be mitigated by incorporating CD40L, an immune-modulator therefore potential molecular adjuvant, into the recombinant adenovirus-based vaccine [Citation72]. Whether this vaccine-associated pathology is related to residual infectious viruses or unbalanced immune responses awaits further investigation. With this in mind, all future MERS-CoV vaccine candidate designs should take extra cautions on safety evaluation.
Furthermore, recombinant-protein-based vaccines are widely pursued. Strategies to solubilize the MERS-CoV S glycoprotein in order to form stable immunogens include forming nanoparticles and using soluble protein truncations. In particular, both nanoparticles formed with full length MERS-CoV S glycoprotein [Citation93,Citation94] and subunit RBD-based vaccines [Citation83–90] have been shown to induce virus neutralizing antibodies and to protect mice when challenged with MERS-CoV. One RBD subunit vaccine also conferred protection in rhesus macaques [Citation91]. This indicates that RBD alone as antigen may be sufficient for protective immunity to develop against the virus. Along with the finding that mAb targeting NTD is able to neutralize MERS-CoV, Lan et al showed that three doses of intramuscularly administered recombinant NTD protein also induced protective immunity against live MERS-CoV in human DPP4 transduced mouse model (Ad5-hDPP4 mice) [Citation92]. More recently, with the structural insights into the spike glycoprotein, Pallesen et al developed a prefusion-stabilized S trimer vaccine by substituting proline residues into the S2 domain [Citation33]. The introduction of proline disfavours the refolding of the linker between HR1 and the central helix, thus preventing the transition of spike into the post-fusion state. This rationally designed antigen, MERS S-2P, was shown to induce broader and more potent neutralizing activity than wild type spike trimer protein [Citation33].
Finally, a prime-boost strategy based on a full-length S glycoprotein DNA vaccine followed by an S1-glycoprotein boost was able to induce virus-neutralizing antibodies and confer protection against the clinical severity of diseases in non-human primate models [Citation49]. Compared with the protein-only regimen, the combination of DNA and protein induced a more functionally diverse antibody repertoire and stronger Th1 immune response. It was suggested that the native S glycoprotein conformation, formed on the cell surface after DNA vaccination, helped induce more diverse antibodies against MERS-CoV.
As summarized in , most of the aforementioned strategies require multiple immunizations which may pose additional logistic hurdles at the end point use. It is unclear whether these immunization strategies were empirically designed or due to relatively poor immunogenicity of candidate vaccines. For practical and compliant purposes, a single immunization with the highest immunogenicity in animals and humans will be preferred.
6. Conclusion
The outbreak of MERS-CoV in Saudi Arabia in 2012 reminded us of the 2003 SARS-CoV outbreak in China. Despite the differences in geographic location, epidemiology and immediate animal reservoirs, these two viruses share remarkable similarity in causing severe respiratory syndrome, leading to high fatality in humans and trigger serious public health concerns. With the advent of modern techniques in virology, immunology and vaccinology, we have gained substantial insights into the biology of MERS-CoV, and its pathogenesis with unprecedented speed and accuracy. As summarized in the current review, tremendous progress has been made in understanding (1) the entry process of MERS-CoV into target cells, (2) the structure and function of S glycoprotein and cellular receptor DPP4 in mediating viral entry, (3) antibody response during natural infection and isolation of broad and potent neutralizing mAbs, and (4) design and development of vaccine candidates using various innovative technologies. However, our progress in translating these discoveries into clinical application has been slow. Only two vaccine candidates and one mAb panel have entered phase I clinical trials for safety. Ironically, no vaccines and treatment strategies have been approved for SARS-CoV infection even after more than a decade of outbreak. We could not imagine how catastrophic it would be should SARS-CoV hit again or MERS-CoV continues to probe and gain strong capacity in transmission to and among humans.
We are facing a difficult predicament when it comes to public health challenges in the new era of emerging and re-emerging infectious diseases. On one hand, the human population is becoming ever mobile and exposed to an increasing number of pathogens. On the other hand, translating basic discoveries into preventative and treatment applications has been exceedingly slow. Among many plausible reasons, a lack of incentives in financial returns perhaps stands the tallest. The deadlock is not just happening to MERS-CoV and SARS-CoV but also to many other infectious pathogens such as Ebola, Marburg, Lassa, highly pathogenic avian influenza, HIV-1, and so on. Fundamental and drastic changes have to be made in the entire research and development system before we can truly prepare and position ourselves ahead of deadly epidemic and pandemic. Only then, can our speed and accuracy in basic discovery be timely translated into clinical and public health needs. The time to act is now.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Kuiken T, Fouchier RA, Schutten M, et al. Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet. 2003 Jul 26;362(9380):263–270. doi:10.1016/S0140-6736(03)13967-0. PubMed PMID: 12892955.
- Zhong NS, Zheng BJ, Li YM, et al. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People’s Republic of China, in February, 2003. Lancet. 2003 Oct 25;362(9393):1353–1358. PubMed PMID: 14585636.
- Hamre D, Procknow JJ. A new virus isolated from the human respiratory tract. Proc Soc Exp Biol Med. 1966 Jan;121(1):190–193. PubMed PMID: 4285768.
- McIntosh K, Dees JH, Becker WB, et al. Recovery in tracheal organ cultures of novel viruses from patients with respiratory disease. Proc Natl Acad Sci USA. 1967 Apr;57(4):933–940. PubMed PMID: 5231356; PubMed Central PMCID: PMCPMC224637.
- van der Hoek L, Pyrc K, Jebbink MF, et al. Identification of a new human coronavirus. Nat Med. 2004 Apr;10(4):368–373. doi:10.1038/nm1024. PubMed PMID: 15034574.
- Woo PC, Lau SK, Chu CM, et al. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J Virol. 2005 Jan;79(2):884–895. doi:10.1128/JVI.79.2.884-895.2005. PubMed PMID: 15613317; PubMed Central PMCID: PMCPMC538593.
- WHO. Summary of probable SARS cases with onset of illness from 1 November 2002 to 31 July 2003 2004 [cited 2019 Mar 6]. Available from: http://www.who.int/csr/sars/country/table2004_04_21/en/
- Zaki AM, van Boheemen S, Bestebroer TM, et al. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012 Nov 8;367(19):1814–1820. doi:10.1056/NEJMoa1211721. PubMed PMID: 23075143.
- WHO. Middle East respiratory syndrome coronavirus (MERS-CoV) 2018 [cited 2019 Mar 26]. Available from: http://www.who.int/emergencies/mers-cov/en/
- Zumla A, Hui DS, Perlman S. Middle East respiratory syndrome. Lancet. 2015 Sep 5;386(9997):995–1007. doi:10.1016/S0140-6736(15)60454-8. PubMed PMID: 26049252; PubMed Central PMCID: PMCPMC4721578.
- Cui J, Li F, Shi ZL. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019 Mar;17(3):181–192. doi:10.1038/s41579-018-0118-9. PubMed PMID: 30531947.
- Woo PC, Lau SK, Li KS, et al. Genetic relatedness of the novel human group C betacoronavirus to Tylonycteris bat coronavirus HKU4 and Pipistrellus bat coronavirus HKU5. Emerg Microbes Infect. 2012 Nov;1(11):e35. doi:10.1038/emi.2012.45. PubMed PMID: 26038405; PubMed Central PMCID: PMCPMC3630921.
- van Boheemen S, de Graaf M, Lauber C, et al. Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. mBio. 2012;3(6):e00473–12.
- Corman VM, Ithete NL, Richards LR, et al. Rooting the phylogenetic tree of middle east respiratory syndrome coronavirus by characterization of a conspecific virus from an African Bat. J Virol. 2014;88(19):11297–11303. doi:10.1128/jvi.01498-14.
- Anthony SJ, Gilardi K, Menachery VD, et al. Further evidence for bats as the evolutionary source of Middle East respiratory syndrome coronavirus. mBio. 2017;8(2). doi:10.1128/mBio.00373-17.
- Wang Q, Qi J, Yuan Y, et al. Bat origins of MERS-CoV supported by bat coronavirus HKU4 usage of human receptor CD26. Cell Host Microbe. 2014;16(3):328–337. doi:10.1016/j.chom.2014.08.009.
- Goldstein SA, Weiss SR. Origins and pathogenesis of Middle East respiratory syndrome-associated coronavirus: recent advances. F1000Res. 2017;6:1628. doi:10.12688/f1000research.11827.1. PubMed PMID: 29026532; PubMed Central PMCID: PMCPMC5583735.
- de Groot RJ, Baker SC, Baric RS, et al. Middle East respiratory syndrome coronavirus (MERS-CoV): announcement of the coronavirus study group. J Virol. 2013 Jul;87(14):7790–7792. doi:10.1128/JVI.01244-13. PubMed PMID: 23678167; PubMed Central PMCID: PMCPMC3700179.
- Qian Z, Dominguez SR, Holmes KV. Role of the spike glycoprotein of human Middle East respiratory syndrome coronavirus (MERS-CoV) in virus entry and syncytia formation. PloS one. 2013;8(10):e76469. doi:10.1371/journal.pone.0076469. PubMed PMID: 24098509; PubMed Central PMCID: PMCPmc3789674; eng.
- Gallagher TM, Buchmeier MJ. Coronavirus spike proteins in viral entry and pathogenesis. Virology. 2001 Jan 20;279(2):371–374. doi:10.1006/viro.2000.0757. PubMed PMID: WOS:000166695000001.
- Masters PS, Pearlman S, et al. Coronaviridae. In: Knipe DM, Howley PM, Cohen JI, editor. Fields virology. Vol. 1. 6th ed. Philadelphia: Lippincott Williams & Wilkins; 2013. p. 825–858.
- Du L, He Y, Zhou Y, et al. The spike protein of SARS-CoV – a target for vaccine and therapeutic development. Nat Rev Microbiol. 2009 Mar;7(3):226–236. doi:10.1038/nrmicro2090. PubMed PMID: 19198616; PubMed Central PMCID: PMCPmc2750777; eng.
- Wang N, Shi X, Jiang L, et al. Structure of MERS-CoV spike receptor-binding domain complexed with human receptor DPP4. Cell Res. 2013;23(8):986–993.
- Lu G, Hu Y, Wang Q, et al. Molecular basis of binding between novel human coronavirus MERS-CoV and its receptor CD26. Nature. 2013: 500(7461):227–231.
- Mou H, Raj VS, van Kuppeveld FJM, et al. The receptor binding domain of the New Middle East respiratory syndrome coronavirus Maps to a 231-residue region in the spike protein that efficiently elicits neutralizing antibodies. J Virol. 2013 Aug;87(16):9379–9383. doi:10.1128/jvi.01277-13. PubMed PMID: WOS:000322535600052.
- Gao J, Lu G, Qi J, et al. Structure of the fusion core and inhibition of fusion by a heptad repeat peptide derived from the S protein of Middle East respiratory syndrome coronavirus. J Virol. 2013;87(24):13134–13140.
- Raj VS, Mou H, Smits SL, et al. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013 Mar 14;495(7440):251–254. doi:10.1038/nature12005. PubMed PMID: WOS:000316652300054.
- Lu L, Liu Q, Zhu Y, et al. Structure-based discovery of Middle East respiratory syndrome coronavirus fusion inhibitor [article]. Nat Commun. 2014 Jan;5:12. doi:10.1038/ncomms4067. PubMed PMID: WOS:000331084200009.
- Wang C, Hua C, Xia S, et al. Combining a fusion inhibitory peptide targeting the MERS-CoV S2 protein HR1 domain and a neutralizing antibody specific for the S1 protein receptor-binding domain (RBD) showed potent Synergism against pseudotyped MERS-CoV with or without mutations in RBD. Viruses. 2019 Jan 6;11(1). doi:10.3390/v11010031. PubMed PMID: 30621343; eng.
- Xia S, Yan L, Xu W, et al. A pan-coronavirus fusion inhibitor targeting the HR1 domain of human coronavirus spike. Sci Adv. 2019 Apr;5(4):eaav4580. doi:10.1126/sciadv.aav4580. PubMed PMID: 30989115; PubMed Central PMCID: PMCPMC6457931.
- Gui M, Song W, Zhou H, et al. Cryo-electron microscopy structures of the SARS-CoV spike glycoprotein reveal a prerequisite conformational state for receptor binding. Cell Res. 2017 Jan;27(1):119–129. doi:10.1038/cr.2016.152. PubMed PMID: 28008928; PubMed Central PMCID: PMCPMC5223232; eng.
- Kirchdoerfer RN, Wang N, Pallesen J, et al. Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis. Sci Rep. 2018 Oct 24;8(1):15701. doi:10.1038/s41598-018-34171-7. PubMed PMID: 30356097; PubMed Central PMCID: PMCPMC6200764; eng.
- Pallesen J, Wang N, Corbett KS, et al. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc Natl Acad Sci USA. 2017 Aug 29;114(35):E7348–E7357. doi:10.1073/pnas.1707304114. PubMed PMID: 28807998; PubMed Central PMCID: PMCPMC5584442.
- Yuan Y, Cao D, Zhang Y, et al. Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains. Nat Commun. 2017 Apr 10;8:15092. doi:10.1038/ncomms15092.
- Song W, Gui M, Wang X, et al. Cryo-EM structure of the SARS coronavirus spike glycoprotein in complex with its host cell receptor ACE2. PLoS Pathog. 2018 Aug;14(8):e1007236. doi:10.1371/journal.ppat.1007236. PubMed PMID: 30102747; PubMed Central PMCID: PMCPMC6107290; eng.
- Li F. Receptor recognition mechanisms of coronaviruses: a decade of structural studies. J Virol. 2015 Feb;89(4):1954–1964. doi:10.1128/JVI.02615-14. PubMed PMID: 25428871; PubMed Central PMCID: PMCPMC4338876.
- Jiang L, Wang N, Zuo T, et al. Potent neutralization of MERS-CoV by human neutralizing monoclonal antibodies to the viral spike glycoprotein. Sci Transl Med. 2014 Apr 30;6(234):234r. a59. doi:10.1126/scitranslmed.3008140. PubMed PMID: 24778414; eng.
- Yu X, Zhang S, Jiang L, et al. Structural basis for the neutralization of MERS-CoV by a human monoclonal antibody MERS-27. Sci Rep. 2015;5:13133. doi:10.1038/srep13133. PubMed PMID: 26281793; PubMed Central PMCID: PMCPmc4539535; eng.
- Zhang S, Zhou P, Wang P, et al. Structural definition of a unique neutralization epitope on the receptor-binding domain of MERS-CoV spike glycoprotein. Cell Rep. 2018 Jul 10;24(2):441–452. doi:10.1016/j.celrep.2018.06.041.
- Tang XC, Agnihothram SS, Jiao Y, et al. Identification of human neutralizing antibodies against MERS-CoV and their role in virus adaptive evolution. Proc Natl Acad Sci USA. 2014 May 13;111(19):E2018–E2026. doi:10.1073/pnas.1402074111. PubMed PMID: 24778221; PubMed Central PMCID: PMCPmc4024880; eng.
- Johnson RF, Bagci U, Keith L, et al. 3B11-N, a monoclonal antibody against MERS-CoV, reduces lung pathology in rhesus monkeys following intratracheal inoculation of MERS-CoV Jordan-n3/2012. Virology. 2016;490:49–58.
- Ying T, Du L, Ju TW, et al. Exceptionally potent neutralization of Middle East respiratory syndrome coronavirus by human monoclonal antibodies. J Virol. 2014 Jul;88(14):7796–7805. doi:10.1128/jvi.00912-14. PubMed PMID: 24789777; PubMed Central PMCID: PMCPmc4097770; eng.
- Ying T, Prabakaran P, Du L, et al. Junctional and allele-specific residues are critical for MERS-CoV neutralization by an exceptionally potent germline-like antibody. Nat Commun. 2015 Sep 15;6(1):8223. doi:10.1038/ncomms9223. PubMed PMID: 26370782; PubMed Central PMCID: PMCPMC4571279.
- van Doremalen N, Falzarano D, Ying T, et al. Efficacy of antibody-based therapies against Middle East respiratory syndrome coronavirus (MERS-CoV) in common marmosets. Antiviral Res. 2017 Apr 5;143:30–37. doi:10.1016/j.antiviral.2017.03.025.
- Houser KV, Gretebeck L, Ying T, et al. Prophylaxis With a Middle East respiratory syndrome coronavirus (MERS-CoV)-specific human monoclonal antibody Protects rabbits from MERS-CoV infection. J Infect Dis. 2016 May 15;213(10):1557–1561. doi:10.1093/infdis/jiw080. PubMed PMID: 26941283; PubMed Central PMCID: PMCPmc4837915; eng.
- Agrawal AS, Ying T, Tao X, et al. Passive transfer of A Germline-like neutralizing human monoclonal antibody Protects transgenic mice against lethal Middle East respiratory syndrome coronavirus infection. Sci Rep. 2016 Aug 19;6:31629. doi:10.1038/srep31629.
- Du L, Zhao G, Yang Y, et al. A conformation-dependent neutralizing monoclonal antibody specifically targeting receptor-binding domain in Middle East respiratory syndrome coronavirus spike protein. J Virol. 2014 Jun;88(12):7045–7053. doi:10.1128/jvi.00433-14. PubMed PMID: 24719424; PubMed Central PMCID: PMCPmc4054355; eng.
- Qiu H, Sun S, Xiao H, et al. Single-dose treatment with a humanized neutralizing antibody affords full protection of a human transgenic mouse model from lethal Middle East respiratory syndrome (MERS)-coronavirus infection. Antiviral Res. 2016 Jun 14;132:141–148. doi:10.1016/j.antiviral.2016.06.003. PubMed PMID: 27312105; Eng.
- Wang L, Shi W, Joyce MG, et al. Evaluation of candidate vaccine approaches for MERS-CoV. Nat Commun. 2015 Jul;6; doi:10.1038/ncomms8712. PubMed PMID: WOS:000358858500018.
- Wang L, Shi W, Chappell JD, et al. Importance of neutralizing monoclonal antibodies targeting multiple antigenic sites on MERS-CoV spike to avoid neutralization escape. J Virol. 2018 Mar 7. doi:10.1128/JVI.02002-17.
- Pascal KE, Coleman CM, Mujica AO, et al. Pre- and postexposure efficacy of fully human antibodies against spike protein in a novel humanized mouse model of MERS-CoV infection. Proc Natl Acad Sci USA. 2015 Jul 14;112(28):8738–8743. doi:10.1073/pnas.1510830112. PubMed PMID: 26124093; PubMed Central PMCID: PMCPmc4507189; eng.
- de Wit E, Feldmann F, Okumura A, et al. Prophylactic and therapeutic efficacy of mAb treatment against MERS-CoV in common marmosets. Antiviral Res. 2018 Aug;156:64–71. doi:10.1016/j.antiviral.2018.06.006.
- Corti D, Zhao J, Pedotti M, et al. Prophylactic and postexposure efficacy of a potent human monoclonal antibody against MERS coronavirus. Proc Natl Acad Sci USA. 2015 Aug 18;112(33):10473–10478. doi:10.1073/pnas.1510199112. PubMed PMID: 26216974; PubMed Central PMCID: PMCPmc4547275; eng.
- de Wit E, Feldmann F, Horne E, et al. Prophylactic efficacy of a human monoclonal antibody against MERS-CoV in the common marmoset. Antiviral Res. 2019 Mar;163:70–74. doi:10.1016/j.antiviral.2019.01.016. PubMed PMID: 30684561.
- Li Y, Wan Y, Liu P, et al. A humanized neutralizing antibody against MERS-CoV targeting the receptor-binding domain of the spike protein. Cell Res. 2015 Nov;25(11):1237–1249. doi:10.1038/cr.2015.113. PubMed PMID: 26391698; PubMed Central PMCID: PMCPmc4650419; eng.
- Chen Z, Bao L, Chen C, et al. Human neutralizing monoclonal antibody inhibition of Middle East respiratory syndrome coronavirus replication in the common marmoset. J Infect Dis. 2017 Jun 15;215(12):1807–1815. doi:10.1093/infdis/jix209.
- Chen Y, Lu S, Jia H, et al. A novel neutralizing monoclonal antibody targeting the N-terminal domain of the MERS-CoV spike protein. Emerg Microbes Infect. 2017 May 24;6(5):e37. doi:10.1038/emi.2017.18.
- Niu P, Zhang S, Zhou P, et al. Ultra-potent human neutralizing antibody Repertoires against MERS-CoV from A recovered patient. J Infect Dis. 2018 May 28. doi:10.1093/infdis/jiy311.
- Niu P, Zhao G, Deng Y, et al. A novel human mAb (MERS-GD27) provides prophylactic and postexposure efficacy in MERS-CoV susceptible mice. Sci China Life Sci. 2018 Oct;61(10):1280–1282. doi:10.1007/s11427-018-9343-8. PubMed PMID: 30091015; eng.
- Zhao G, He L, Sun S, et al. A novel nanobody targeting Middle East respiratory syndrome coronavirus (MERS-CoV) receptor-binding domain has potent cross-neutralizing activity and protective efficacy against MERS-CoV. J Virol. 2018 Sep 15;92(18). doi:10.1128/JVI.00837-18.
- Stalin Raj V, Okba NMA, Gutierrez-Alvarez J, et al. Chimeric camel/human heavy-chain antibodies protect against MERS-CoV infection. Sci Adv. 2018 Aug;4(8):eaas9667. doi:10.1126/sciadv.aas9667. PubMed PMID: 30101189; PubMed Central PMCID: PMCPMC6082650.
- Agrawal AS, Tao X, Algaissi A, et al. Immunization with inactivated Middle East respiratory syndrome coronavirus vaccine leads to lung immunopathology on challenge with live virus. Hum Vaccin Immunother. 2016 Sep;12(9):2351–2356. doi:10.1080/21645515.2016.1177688. PubMed PMID: 27269431; PubMed Central PMCID: PMCPMC5027702.
- Deng Y, Lan J, Bao L, et al. Enhanced protection in mice induced by immunization with inactivated whole viruses compare to spike protein of Middle East respiratory syndrome coronavirus. Emerg Microbes Infect. 2018 Apr 4;7(1):60. doi:10.1038/s41426-018-0056-7.
- Wang C, Zheng X, Gai W, et al. MERS-CoV virus-like particles produced in insect cells induce specific humoural and cellular imminity in rhesus macaques. Oncotarget. 2017 Feb 21;8(8):12686–12694. doi:10.18632/oncotarget.8475. PubMed PMID: 27050368; PubMed Central PMCID: PMCPMC5355045.
- Song F, Fux R, Provacia LB, et al. Middle East respiratory syndrome coronavirus spike protein delivered by modified Vaccinia virus Ankara efficiently induces virus-neutralizing antibodies. J Virol. 2013 Nov;87(21):11950–11954. doi:10.1128/jvi.01672-13. PubMed PMID: WOS:000325863400060.
- Volz A, Kupke A, Song F, et al. Protective efficacy of recombinant modified Vaccinia virus Ankara delivering Middle East respiratory syndrome coronavirus spike glycoprotein. J Virol. 2015 Aug;89(16):8651–8656. doi:10.1128/jvi.00614-15. PubMed PMID: WOS:000358278200047.
- Haagmans BL, van den Brand JM, Raj VS, et al. An orthopoxvirus-based vaccine reduces virus excretion after MERS-CoV infection in dromedary camels. Science. 2016 Jan 1;351(6268):77–81. doi:10.1126/science.aad1283. PubMed PMID: 26678878; eng.
- Kim E, Okada K, Kenniston T, et al. Immunogenicity of an adenoviral-based Middle East respiratory syndrome coronavirus vaccine in BALB/c mice. Vaccine. 2014 Oct 14;32(45):5975–5982. doi:10.1016/j.vaccine.2014.08.058. PubMed PMID: WOS:000343629900014.
- Guo X, Deng Y, Chen H, et al. Systemic and mucosal immunity in mice elicited by a single immunization with human adenovirus type 5 or 41 vector-based vaccines carrying the spike protein of Middle East respiratory syndrome coronavirus. Immunology. 2015 Aug;145(4):476–484. doi:10.1111/imm.12462. PubMed PMID: WOS:000357854300003.
- Munster VJ, Wells D, Lambe T, et al. Protective efficacy of a novel simian adenovirus vaccine against lethal MERS-CoV challenge in a transgenic human DPP4 mouse model. NPJ Vaccines. 2017 Oct 16;2:28. doi:10.1038/s41541-017-0029-1.
- Alharbi NK, Padron-Regalado E, Thompson CP, et al. Chadox1 and MVA based vaccine candidates against MERS-CoV elicit neutralising antibodies and cellular immune responses in mice. Vaccine. 2017 Jun 27;35(30):3780–3788. doi:10.1016/j.vaccine.2017.05.032.
- Hashem AM, Algaissi A, Agrawal A, et al. A highly immunogenic, protective and safe adenovirus-based vaccine expressing MERS-CoV S1-CD40L fusion protein in transgenic human DPP4 mouse model. J Infect Dis. 2019 Mar 26. doi:10.1093/infdis/jiz137. PubMed PMID: 30911758.
- Jia W, Channappanavar R, Zhang C, et al. Single intranasal immunization with chimpanzee adenovirus-based vaccine induces sustained and protective immunity against MERS-CoV infection. Emerg Microbes Infect. 2019 May 28;8(1):760–772. doi:10.1080/22221751.2019.1620083. PubMed PMID: 31130102.
- Malczyk AH, Kupke A, Prufer S, et al. A highly Immunogenic and protective Middle East respiratory syndrome coronavirus vaccine based on a recombinant measles virus vaccine platform. J Virol. 2015 Nov;89(22):11654–11667. doi:10.1128/jvi.01815-15. PubMed PMID: 26355094; PubMed Central PMCID: PMCPmc4645655; eng.
- Bodmer BS, Fiedler AH, Hanauer JRH, et al. Live-attenuated bivalent measles virus-derived vaccines targeting Middle East respiratory syndrome coronavirus induce robust and multifunctional T cell responses against both viruses in an appropriate mouse model. Virology. 2018 Jun 11;521:99–107. doi:10.1016/j.virol.2018.05.028.
- Zhao J, Li K, Wohlford-Lenane C, et al. Rapid generation of a mouse model for Middle East respiratory syndrome. Proc Natl Acad Sci USA. 2014 Apr 1;111(13):4970–4975. doi:10.1073/pnas.1323279111. PubMed PMID: 24599590; PubMed Central PMCID: PMCPMC3977243.
- Li K, Wohlford-Lenane C, Perlman S, et al. Middle East respiratory syndrome coronavirus Causes multiple Organ Damage and lethal disease in mice transgenic for human dipeptidyl peptidase 4. J Infect Dis. 2016 Mar 1;213(5):712–722. doi:10.1093/infdis/jiv499. PubMed PMID: 26486634; PubMed Central PMCID: PMCPMC4747621.
- Liu R, Wang J, Shao Y, et al. A recombinant VSV-vectored MERS-CoV vaccine induces neutralizing antibody and T cell responses in rhesus monkeys after single dose immunization. Antiviral Res. 2018 Feb;150:30–38. doi:10.1016/j.antiviral.2017.12.007. PubMed PMID: 29246504.
- Wirblich C, Coleman CM, Kurup D, et al. One-health: a safe, efficient, dual-use vaccine for humans and animals against Middle East respiratory syndrome coronavirus and rabies virus. J Virol. 2017 Jan 15;91(2). doi:10.1128/JVI.02040-16.
- Jung S-Y, Kang KW, Lee E-Y, et al. Heterologous prime-boost vaccination with adenoviral vector and protein nanoparticles induces both Th1 and Th2 responses against Middle East respiratory syndrome coronavirus. Vaccine. 2018 May 5;36(24):3468–3476. doi:10.1016/j.vaccine.2018.04.082.
- Muthumani K, Falzarano D, Reuschel EL, et al. A synthetic consensus anti-spike protein DNA vaccine induces protective immunity against Middle East respiratory syndrome coronavirus in nonhuman primates. Sci Transl Med. 2015 Aug 19;7(301):301ra132. doi:10.1126/scitranslmed.aac7462. PubMed PMID: 26290414; PubMed Central PMCID: PMCPmc4573558; eng.
- Chi H, Zheng X, Wang X, et al. DNA vaccine encoding Middle East respiratory syndrome coronavirus S1 protein induces protective immune responses in mice. Vaccine. 2017 Apr 11;35(16):2069–2075. doi:10.1016/j.vaccine.2017.02.063.
- Du L, Kou Z, Ma C, et al. A Truncated receptor-binding domain of MERS-CoV spike protein potently Inhibits MERS-CoV infection and induces strong neutralizing antibody responses: Implication for developing therapeutics and vaccines. PloS one. 2013 Dec 4;8(12). doi:10.1371/journal.pone.0081587. PubMed PMID: WOS:000327949300098.
- Ma C, Wang L, Tao X, et al. Searching for an ideal vaccine candidate among different MERS coronavirus receptor-binding fragments – the importance of immunofocusing in subunit vaccine design. Vaccine. 2014 Oct 21;32(46):6170–6176. doi:10.1016/j.vaccine.2014.08.086.
- Zhang N, Channappanavar R, Ma C, et al. Identification of an ideal adjuvant for receptor-binding domain-based subunit vaccines against Middle East respiratory syndrome coronavirus. Cell Mol Immunol. 2015 Feb 2. doi:10.1038/cmi.2015.03. PubMed PMID: 25640653; Eng.
- Tang J, Zhang N, Tao X, et al. Optimization of antigen dose for a receptor-binding domain-based subunit vaccine against MERS coronavirus. Hum Vaccin Immunother. 2015 May 4;11(5):1244–1250. doi:10.1080/21645515.2015.1021527. PubMed PMID: WOS:000355117100035.
- Wang Y, Tai W, Yang J, et al. Receptor-binding domain of MERS-CoV with optimal immunogen dosage and immunization interval protects human transgenic mice from MERS-CoV infection. Hum Vaccin Immunother. 2017 Jul 3;13(7):1615–1624. doi:10.1080/21645515.2017.1296994.
- Tai W, Zhao G, Sun S, et al. A recombinant receptor-binding domain of MERS-CoV in trimeric form protects human dipeptidyl peptidase 4 (hDPP4) transgenic mice from MERS-CoV infection. Virology. 2016 Oct 15;499:375–382. doi:10.1016/j.virol.2016.10.005.
- Ma C, Li Y, Wang L, et al. Intranasal vaccination with recombinant receptor-binding domain of MERS-CoV spike protein induces much stronger local mucosal immune responses than subcutaneous immunization: Implication for designing novel mucosal MERS vaccines. Vaccine. 2014 Apr 11;32(18):2100–2108. doi:10.1016/j.vaccine.2014.02.004. PubMed PMID: WOS:000334980800014.
- Lan J, Deng Y, Chen H, et al. Tailoring subunit vaccine immunity with adjuvant combinations and delivery routes using the Middle East respiratory coronavirus (MERS-CoV) receptor-binding domain as an antigen. PloS one. 2014;9(11):e112602. doi:10.1371/journal.pone.0112602. PubMed PMID: 25405618; PubMed Central PMCID: PMCPmc4236105; eng.
- Lan J, Yao Y, Deng Y, et al. Recombinant receptor binding domain protein induces Partial protective Immunity in rhesus macaques against Middle East respiratory syndrome coronavirus challenge. EBioMed. 2015 Oct;2(10):1438–1446. doi:10.1016/j.ebiom.2015.08.031. PubMed PMID: 26629538; PubMed Central PMCID: PMCPmc4634622; eng.
- Lan J, Yao Y, Deng Y, et al. The recombinant N-terminal domain of spike proteins is a potential vaccine against Middle East respiratory syndrome coronavirus (MERS-CoV) infection. Vaccine. 2017 Jan 3;35(1):10–18. doi:10.1016/j.vaccine.2016.11.064. PubMed PMID: 27899228.
- Coleman CM, Liu YV, Mu H, et al. Purified coronavirus spike protein nanoparticles induce coronavirus neutralizing antibodies in mice. Vaccine. 2014 May 30;32(26):3169–3174. doi:10.1016/j.vaccine.2014.04.016. PubMed PMID: WOS:000336872500009.
- Coleman CM, Venkataraman T, Liu YV, et al. MERS-CoV spike nanoparticles protect mice from MERS-CoV infection. Vaccine. 2017 Mar 14;35(12):1586–1589. doi:10.1016/j.vaccine.2017.02.012.
- McCoy K, Tatsis N, Korioth-Schmitz B, et al. Effect of preexisting immunity to adenovirus human serotype 5 antigens on the immune responses of nonhuman primates to vaccine regimens based on human- or chimpanzee-derived adenovirus vectors. J Virol. 2007 Jun;81(12):6594–6604. doi:10.1128/JVI.02497-06. PubMed PMID: 17428852; PubMed Central PMCID: PMCPMC1900096.
- Mast TC, Kierstead L, Gupta SB, et al. International epidemiology of human pre-existing adenovirus (Ad) type-5, type-6, type-26 and type-36 neutralizing antibodies: correlates of high Ad5 titers and implications for potential HIV vaccine trials. Vaccine. 2010 Jan 22;28(4):950–957. doi:10.1016/j.vaccine.2009.10.145. PubMed PMID: 19925902.
- Farina SF, Gao GP, Xiang ZQ, et al. Replication-defective vector based on a chimpanzee adenovirus. J Virol. 2001 Dec;75(23):11603–11613. doi:10.1128/JVI.75.23.11603-11613.2001. PubMed PMID: 11689642; PubMed Central PMCID: PMCPMC114747.