ABSTRACT
The NF-κB pathway regulates cell physiology under stress conditions. We have recently described a novel NF-κB regulatory mechanism, by which SIRT6 induces cysteine monoubiquitination of the methyltransferase SUV39H1. This causes SUV39H1 dissociation from the gene encoding the NF-κB inhibitor IκBα, increasing its expression and leading to NF-κB pathway inactivation.
The transcription factor NF-κB plays a key role in stress response due to environmental stimuli by regulating genes involved in proliferation, apoptosis, differentiation, inflammation and immune system regulation. In non-stress conditions, NF-κB is sequestered in the cytoplasm by its repressor IκBα (encoded by the gene Nfkb1a). Under stress conditions, IκBα is degraded, allowing the translocation of NF-κB transcription factors to the nucleus, and subsequently activating the transcription of key genes.Citation1 Given the importance of these target genes and their tissue-specific nature, NF-κB pathway activation is tightly regulated through several mechanisms. One of them involves the NAD+-dependent deacetylase and ADP-ribosyltransferase SIRT6, a member of the sirtuin family involved in stress-related regulation of genome stability, inflammation and metabolism.Citation2 After the pathway activation, SIRT6 is recruited to the promoter regions of NF-κB target genes where it promotes gene silencing by deacetylating acetylated lysine 9 of histone H33 ().
Figure 1. Dual Regulation of NF-κB signaling by SIRT6. SIRT6 regulates NF-κB pathway hyperactivation in two ways: First, SIRT6 deacetylates H3K9 in promoters of NF-κB target genes and, subsequently, induces specific gene silencing; Second, SIRT6 promotes suppressor of variegation 3–9 homolog 1 (SUV39H1) cysteine monoubiquitination by recruiting and regulating the E3-ubiquitin ligase S-phase associate protein kinase 2 (SKP2) inducing the dissociation of SUV39H1 from the promoter of Nfkb1a, the gene encoding the NF-κB negative regulator, IκBα. This in turn inhibits NF-κB nuclear localization and promotes a general downregulation of the pathway. Ac: acetyl. Ub: ubiquitin.
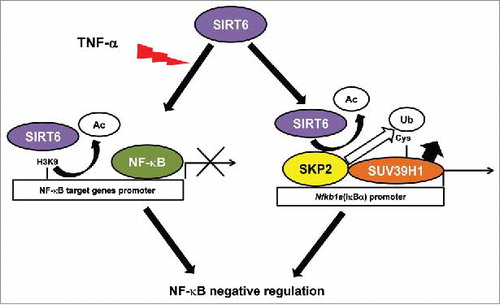
In our recent work,Citation4 we have described a new mechanism of NF-κB regulation mediated by SIRT6. SIRT6 promotes the expression of the NF-κB repressor, IκBα, by inducing cysteine monoubiquitination of the histone methyltransferase Suppressor of variegation 3–9 homolog 1 (SUV39H1), which promotes SUV39H1 dissociation from the Nfkb1a promoter (). SUV39H1 is a major regulator of gene silencing, genome organization and heterochromatin formation through catalysis of tri-methylation of histone H3 at lysine 9.Citation5 Dissociation of SUV39H1 enables Nfkb1a to be transcribed, increasing IκBα protein levels, which restores NF-κB cytoplasmic re-localization and abrogates NF-κB pathway activation. This mechanism is cell-type specific, which is in concordance with the cell-type specific behavior and regulatory mechanisms associated with the NF-κB pathway.Citation6 Interestingly, we have previously reported that the stability of SUV39H1 can also be regulated by another sirtuin, SIRT1, upon oxidative stress.Citation7 However, despite the similarity between SIRT1 and SIRT6, their regulation of SUV39H1 varies as they respond to different types of stress, bind to different SUV39H1 domains, and have distinct mechanisms.
Ubiquitination is a common post-translational protein modification involved in many functions from proteasomal degradation to cell signaling. It involves the addition of one or more 8.5kDa ubiquitin molecules to the target protein (mono- and polyubiquitination, respectively). Although these modifications usually target lysine residues and involve the formation of an isopeptide bond (canonical ubiquitination), they can also occur on other residues such as cysteines, threonines, serines or tyrosines through thioester or hydroxyesther bonds (non-canonical ubiquitination).Citation8 To date, we know very few cases of non-canonical ubiquitination, described in major histocompatibility complex I (MHC-I) proteins, peroxisomal proteins and in the proneural transcription factor NEUROG2. These types of ubiquitination are rarely reported, perhaps because they occur at low frequencies or due to the higher lability of these non-canonical bonds. In our study, mass spectrometry analysis of SUV39H1 monoubiquitination induced by SIRT6 identified four cysteines and one serine as targets. Three of the identified cysteines were part of the nine conserved cysteines in the SUV39H1 PRE-SET domain, a domain previously suggested to participate in SUV39H1 binding to chromatin. This is in agreement with our findings as both the mutation of the conserved ubiquitinated cysteines and the activation of the NF-κB pathway, with the cytokine Tumor necrosis factor (TNF-α), promotes SUV39H1 dissociation from the Nfkb1a promoter. However, further structural studies are required to explain how cysteine monoubiquitination affects the PRE-SET domain and interferes with SUV39H1 chromatin binding ability. Interestingly, the other ubiquitinated cysteine and serine residues are present in two regions that have been also associated with SUV39H1 chromatin binding: the N-terminal region (serine) and the methyl-lysine binding specific chromodomain (cysteine). These findings strongly suggest that target of SUV39H1 monoubiquitination is not a specific residue but rather the chromatin binding ability of SUV39H1.
We identified S-phase associate protein kinase 2 (SKP2), a F-box E3 ubiquitin ligase, as the enzyme responsible for SUV39H1 cysteine monoubiquitination. SKP2 is considered an oncogene because it promotes cell proliferation.Citation9 SKP2 is also part of the cell cycle regulating SCF complex, and regulates cell cycle progression by catalyzing canonical polyubiquitination and subsequent proteasomal degradation of checkpoints regulators, such as Cyclin-dependent kinase inhibitors 1, 1B, 1C (known as p21, p27 and p57, respectively) and Cyclin E. Our work is the first report that describes SKP2 as a non-canonical E3 monoubiquitin ligase suggesting that it might be involved in other unknown regulatory mechanisms. We demonstrate that SIRT6 recruits SKP2 to the Nfkb1a promoter allowing monoubiquitination of SUV39H1. Moreover, SIRT6 ensures SKP2 protein stability through its deacetylation. SKP2 levels are regulated during the cell cycle through phosphorylation of Nuclear localization signal residues S72 and S75 by Protein kinase B (known as AKT) and Casein Kinase I (CKI), respectively. Phosphorylation of both sites prevents the proteasomal degradation of SKP2.Citation10 SIRT6 deacetylates the nearby residues K73 and K77 which in turn promotes S72/S75 phosphorylation and SKP2 stability. It is still unclear whether SIRT6 regulation of SKP2 is cell–cycle dependent, however, the fact that SUV39h1 monoubiquitination is regulated during S-phase suggests that this may be the case.
Our model suggests that SIRT6 prevents overactivation of the NF-κB pathway, not only by deacetylating H3K9 at the promoter of NF-κB target genes, but also by activating the NF-κB repressor, IκBα (). We hypothesize that this double regulatory mechanism is an efficient way to ensure fast, dynamic and specific control of NF-κB pathway activation. Our work also supports that non-canonical ubiquitination plays a relevant role in mammalian cell physiology. Future studies should define the range and functional relevance of these modifications, which we anticipate will change our perspective about protein ubiquitination.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Additional information
Funding
References
- Hoesel B, Schmid JA. The complexity of NF-κB signaling in inflammation and cancer. Mol Cancer. 2013;12:86. doi:10.1186/1476-4598-12-86. PMID:23915189
- Kugel S, Mostoslavsky R. Chromatin and beyond : the multitasking roles for SIRT6. Trends Biochem Sci. 2014;39:72–81. doi:10.1016/j.tibs.2013.12.002. PMID:24438746
- Kawahara TLA, Michishita E, Adler AS, Damian M, Berber E, Lin M, McCord RA, Ongaigui KC, Boxer LD, Chang HY, et al. SirT6 links histone H3 lysine 9 deacetylation to NF-kB-dependent gene expression and organismal lifespan. Cell. 2009;136:62–74. doi:10.1016/j.cell.2008.10.052. PMID:19135889.
- Santos-Barriopedro I, Bosch-Presegué L, Marazuela-Duque A, de la, Torre C, Colomer C, Vazquez BN, Fuhrmann T, Martínez-Pastor B, Lu W, Braun T, et al. SIRT6-dependent cysteine monoubiquitination in the PRE-SET domain of Suv39h1 regulates the NF-κB pathway. Nat Commun. 2018;9:101. doi:10.1038/s41467-017-02586-x. PMID:29317652
- Rao VK, Pal A, Taneja R. A drive in SUVs: From development to disease. Epigenetics. 2017;12:177–86. doi:10.1080/15592294.2017.1281502. PMID:28106510
- Sen R, Smale ST. Selectivity of the NF-κB response. Cold Spring Harb Perspect Biol. 2010;2:a000257. doi: 10.1101/cshperspect.a000257. PMID: 20452937.
- Bosch-Presegué L, Raurell-Vila H, Marazuela-Duque A, Kane-Goldsmith N, Valle A, Oliver J, Serrano L, Vaquero A. Stabilization of Suv39H1 by SirT1 is part of oxidative stress response and ensures genome protection. Mol Cell. 2011;42:210–23. doi:10.1016/j.molcel.2011.02.034. PMID:21504832
- McDowell GS, Philpott A. Non-canonical ubiquitylation: mechanisms and consequences. Int J Biochem Cell Biol. 2013;45:1833–42. doi:10.1016/j.biocel.2013.05.026. PMID:23732108
- Chan CH, Li CF, Yang WL, Gao Y, Lee SW, Feng Z, Huang HY, Tsai KK, Flores LG, Shao Y, et al. The Skp2-SCF E3 ligase regulates Akt ubiquitination, glycolysis, herceptin sensitivity, and tumorigenesis. Cell. 2012; 149:1098–111. doi: 10.1016/j.cell.2012.02.065. PMID: 22632973.
- Ecker K, Hengst L. Skp2: caught in the Akt. Nat Cell Biol. 2009;11:377–9. doi:10.1038/ncb0409-377. PMID:19337320
