ABSTRACT
Rhabdomyosarcoma (RMS) histologically resembles developing skeletal muscle and is thought to solely originate from a differentiation block in muscle progenitors. We demonstrate that RMS can arise from endothelial progenitor cells following reprogramming and myogenic transdifferentiation. These results highlight how tumors with identical morphological features can arise from different cell types and offer insight into RMS formation in non-myogenic tissue.
Rhabdomyosarcoma (RMS) is the most common soft tissue sarcoma occurring in children. Despite aggressive treatment and intensive research, clinical outcomes for high-risk patients have not significantly improved over the past three decades highlighting the need to uncover the molecular underpinnings of RMS. RMS is subdivided into two major classes, fusion-positive and fusion-negative, based on the presence or absence of the PAX3-FOXO1 or PAX7-FOXO1 gene fusions.Citation1 RMS resembles the histology and gene expression of developing skeletal muscle and had been thought to originate exclusively from skeletal muscle progenitors.Citation2 However, an exclusively myogenic origin does not account for the clinical occurrence of RMS in tissues devoid of skeletal muscle such as the bladder, prostate, salivary gland and biliary tree. Although approximately 40% of RMS occurs in the head and neck, RMS can occur throughout the body and tumor location is a key feature of staging and thus prognosis. It remains unknown what drives RMS in particular locations and the mechanism by which location impacts survival.
Previously, we described a genetically engineered mouse model of fusion-negative RMS (FN-RMS) resulting from Hedgehog signaling through expression of a conditional, constitutively active Smoothened allele, SmoM2, driven by Cre recombinase expressed from the adipose protein 2 (aP2) promoter. In our model, aP2-Cre;SmoM2 mutant mice develop aggressive tumors in the head and neck that display the histologic and molecular characteristics of human FN-RMS.Citation3 Now, we leveraged this mouse model and used genetic fate mapping to interrogate the cell of origin of FN-RMS.Citation4
The most intriguing aspect of our model was FN-RMS developing from cells expressing aP2-Cre. Previously, aP2 expression was thought to be adipose specific but broader spectrum of expression is now evident.Citation5 We sought to identify the aP2-Cre expressing cell type that resulted in FN-RMS. We illustrated that in both the absence and presence of oncogenic SmoM2 skeletal muscle does not derive from aP2-Cre expressing cells. We further demonstrated that aP2-Cre is not expressed during muscle stem cell (satellite cell) specification, activation or resulting differentiation thus confirming that FN-RMS originates from a non-myogenic origin in the aP2-Cre;SmoM2 model. Instead, we found that aP2-Cre labels cells nestled within the interstitial space between skeletal muscle fibers.
Our gene fate mapping with fluorescent labeled cells allowed us to visualize the early stages of tumorigenesis as well as to develop a fluorescent-activated cell sorting (FACS) method to purify tumor cells from the heterogeneous solid tumor cell population. The early onset of tumors in our model suggested that transformation likely occurred during embryogenesis. By analyzing mouse embryos, we found aP2-Cre labeled cells proliferated and expanded within the head and neck muscle interstitium as early as embryonic day E15.5. FACS isolation and gene expression analysis of aP2-labeled cells from the neck skeletal muscle of non-tumor bearing adult mice illustrated that these aP2-labeled skeletal muscle interstitial cells were endothelial cells.
Mature endothelial cells labeled by aP2-Cre within the muscle interstitium of adult mice express the SmoM2 transgene but do not exhibit an active Hedgehog signaling gene expression signature. Therefore, mature endothelial cells without an active hedgehog pathway are not the origin of FN-RMS in the aP2-Cre;SmoM2 model. The aP2-Cre labeled embryonic cell expansions do not express PECAM1 and instead become Myogenic differentiation 1 (MYOD1) positive by embryonic day E18.5, thus suggesting that oncogenic SmoM2 drives myogenic transdifferentiation by reprogramming endothelial progenitor cells into FN-RMS. The activation of the hedgehog pathway was pivotal to FN-RMS formation as activation of oncogenic Kirsten rat sarcoma viral oncogene homolog (KRASG12D) and deletion of Cyclin dependent kinase inhibitor 2a (Cdkn2a) with aP2-Cre resulted in angiosarcoma, an endothelial tumor, not FN-RMS. As well, deletion of Dicer1 with aP2-Cre results angiosarcoma.Citation6 These results illustrate how the cell of origin and specific transforming events intersect and cooperate to impact the tumor cell identity.
Gene expression analysis of purified tumor cells gave insight into the mechanism of myogenic transdifferentiation. Gene ontology analysis found a number of tissue morphogenesis genes to be significantly enriched in isolated tumor cells. Among these were T-box1 (TBX1), Paired-like homeodomain transcription factor 2 (PITX2), Transcription factor 21 (TCF21) and Musculin (MSC), transcription factors involved in the specification of head and neck muscle progenitors acting upstream of MYOD1.Citation7 Interestingly, Paired box 3 (PAX3), which specifies muscle progenitor cells in the trunk and limb, was not expressed. TBX1 is expressed in response to Sonic hedgehog pathway activationCitation8 and drives MYOD1 expression in head and neck muscle progenitor cells following their commitment to the myogenic lineage. Consistent with SmoM2 driving rhabdomyosarcomagenesis through activation of myogenic specification factors, we find Tbx1 expressed in embryonic expansions of aP2-Cre labeled cells at embryonic day E15.5 prior to MYOD1 expression at embryonic day E18.5.
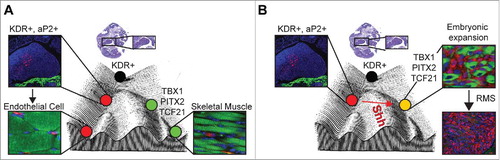
We showed that endothelium and head and neck skeletal muscle arise from multipotent Kinase insert domain receptor (KDR, also known as FLK1 and VEGFR2) expressing progenitor cells and that aP2-Cre is expressed in these KDR expressing progenitor cells following their commitment to the endothelial cell lineage. Oncogenic SmoM2 expression drives the embryonic expansion, transformation, and reprogramming of these aP2-Cre expressing cells in the head and neck during tumorigenesis (). These findings demonstrate that cell fate reprogramming resulting in transdifferentiation can drive tumorigenesis in pediatric sarcoma and illustrate how normal development programs are hijacked to drive tumor location. Our genetic fate mapping identified aP2-labeled endothelial cells within the skeletal muscle interstitium throughout the mouse; however, tumors only develop in the head and neck in our model. This work highlights the interplay between the drivers of transformation and the cellular context in which transformation occurs both providing contributions to tumorigenesis. The cell of origin of tumors are typically inferred from characteristics of the tumor cell. Our results highlight why such assumptions can be dangerous.
Figure 1. Role for endothelial progenitor reprogramming by Hedgehog in rhabdomyosarcoma oncogenesis. A) Model of endothelial cell and skeletal muscle development from Kinase insert domain receptor positive (KDR+) progenitors in the pharyngeal mesoderm on a Waddington epigenetic landscape. B) Proposed model of fusion-negative rhabdomyosarcoma tumorigenesis. Constitutive activation of Hedgehog signaling (Shh) by expression of constitutively active oncogenic SMOOTHENED (SmoM2) in aP2-Cre expressing endothelial progenitor cells results in their embryonic expansion, reprogramming and myogenic transdifferentiation into fusion-negative rhabdomyosarcoma. Immunostaining for Tomato (Red), Myosin Heavy Chain (Green) and DAPI (Blue). Abbreviations in figure as follows: adipose protein 2 (aP2), T-box 1 (TBX1), Paired-like homeodomain transcription factor 2 (PITX2), Transcription factor 21 (TCF21), rhabdomyosarcoma (RMS). Adapted from.Citation9
Disclosure statement
The authors declare no conflicts of interest.
Additional information
Funding
References
- Borinstein SC, Steppan D, Hayashi M, Loeb DM, Isakoff MS, Binitie O, Brohl AS, Bridge JA, Stavas M, Shinohara ET, et al. Consensus and controversies regarding the treatment of rhabdomyosarcoma. Pediatr Blood Cancer. 2018;65. doi:10.1002/pbc.26809.
- Kashi VP, Hatley ME, Galindo RL. Probing for a deeper understanding of rhabdomyosarcoma: insights from complementary model systems. Nat Rev Cancer. 2015;15:426–39. doi:10.1038/nrc3961. PMID:26105539.
- Hatley ME, Tang W, Garcia MR, Finkelstein D, Millay DP, Liu N et al. A mouse model of rhabdomyosarcoma originating from the adipocyte lineage. Cancer cell 2012; 22: 536–46. doi:10.1016/j.ccr.2012.09.004. PMID:23079662.
- Drummond CJ, Hanna JA, Garcia MR, Devine DJ, Heyrana AJ, Finkelstein D, Rehg JE, Hatley ME. Hedgehog Pathway Drives Fusion-Negative Rhabdomyosarcoma Initiated From Non-myogenic Endothelial Progenitors. Cancer Cell. 2018;33:108–124.e105. doi:10.1016/j.ccell.2017.12.001. PMID:29316425.
- Lee KY, Russell SJ, Ussar S, Boucher J, Vernochet C, Mori MA, Smyth G, Rourk M, Cederquist C, Rosen ED, et al. Lessons on conditional gene targeting in mouse adipose tissue. Diabetes. 2013;62:864–74. doi:10.2337/db12-1089. PMID:23321074.
- Hanna JA, Drummond CJ, Garcia MR, Go JC, Finkelstein D, Rehg JE, Hatley ME. Biallelic Dicer1 loss mediated by aP2-Cre drives angiosarcoma. Cancer Res. 2017. 77:6109–6118 doi:10.1158/0008-5472.CAN-17-1262. PMID:28916654.
- Buckingham M. Gene regulatory networks and cell lineages that underlie the formation of skeletal muscle. Proc Natl Acad Sci U S A. 2017. 114:5830–5837 doi:10.1073/pnas.1610605114.
- Garg V, Yamagishi C, Hu T, Kathiriya IS, Yamagishi H, Srivastava D. Tbx1, a DiGeorge syndrome candidate gene, is regulated by sonic hedgehog during pharyngeal arch development. Dev Biol 2001; 235: 62–73. doi:10.1006/dbio.2001.0283. PMID:11412027.
- Jones KB. What's in a Name? Cell Fate Reprogramming in Sarcomagenesis. Cancer Cell. 2018;33:5–7. doi:10.1016/j.ccell.2017.12.005. PMID:29316432.
