Abstract
The complete mitogenome of Anodonta cygnea is 15,613 bp long. This compact, circular molecule contains the set of 37 genes, typical for invertebrate mitogenomes, in the same order and orientation as in maternally inherited genomes of other bivalves from the same subfamily. There are only two unassigned regions longer than 200 bp (266 bp and 274 bp) and no indication of any supranumerary open reading frames.
Keywords:
Anodonta cygnea (Linnaeus, 1758) is a freshwater mussel from the family Unionidae, distributed in Eurasian waters. The family is several hundred species rich, but most of them are found in North America. They are usually gonochoristic, with the presence of two distinct mitochondrial lineages (M and F), inherited under DUI system (Skibinski et al. Citation1994; Zouros et al. Citation1994). This system has been faithfully operating in freshwater mussels for a long time, leading to extreme divergence of the two mitogenomes (Hoeh et al. Citation2002). Gender-specific anonymous open reading frames (FORF and MORF) have been described in both mitogenomes (Doucet-Beaupré et al. Citation2010). The few species with secondary hermaphroditism were described, and in case of North American mussels, these always lost the divergent, paternally inherited mitogenome. There were also substantial structural changes in the FORF (now denoted HORF) (Breton et al. Citation2011).
Here we announce, for the first time, the mitogenome of a European hermaphroditic species from the same family. We were unable to find a distinct paternally inherited mitogenome in sperm of this species so we assume the announced mitogenome to be the only one present.
The sample was taken in July 2009 from a pond in Hamrzysko village, central Poland. Identification down to species level was based on diagnostic morphological characters (Piechocki and Dyduch-Falniowska Citation1993). The specimen is stored under voucher number 328 in the local collection at University of Szczecin. The taxonomic identity was confirmed by comparison of the barcoding cox1 sequence with the references (Bogan and Roe Citation2008).
The sequencing strategy followed the previously published three-step protocol (Soroka and Burzyński Citation2010). Two parts of the mitogenome were amplified with universal primers and sequenced. Species-specific long-range primers were used to amplify the rest of the mitogenome. The LR-PCR products were sequenced by primer walking. The complete mitogenome was assembled in gap4 from Staden package (Staden et al. Citation2001). Annotations followed the established pipeline (Zbawicka et al. Citation2007) and were manually curated by comparison with the mitogenome of A. anatina (Soroka and Burzyński Citation2015).
The sequence has been deposited in GenBank under accession number MG385135. Comparative phylogenetic analysis was performed (). The protein sequences encoded by the mitogenome differ from the closest relative (A. anatina F mitogenome) by approximately 10% (average p-distance, calculated in MEGA7 (Kumar et al. Citation2016)). No additional ORFs could be identified. In particular, the region containing FORF in A. anatina F mitogenome and HORF in Utterbackia imbecillis and Lasmigona compressa mitogenomes is much shorter and does not contain any ORF of appreciable length in A. cygnea.
Figure 1. Phylogenetic tree showing the relationship of the announced mitogenome (in boldface) with the 23 closest relatives identified by BLAST search in nr database. It places A. cygnea as a sister taxon to A. anatina. The “latin name” fields of GenBank records were used to identify sequences, but several of these may be incorrect. In particular, the species related to Sinanodonta woodiana should most likely not be classified as Anodonta, likewise the three Lamprotula species should also follow different naming convention, most likely that presented recently by Lopes-Lima et al. (Citation2017). The following records were used: Aculamprotula tientsinensis KR873102 (Wu et al. Citation2016), Anodonta anatina KF030964 (Soroka and Burzyński Citation2015), Anodonta arcaeformis KF667530 (An et al. Citation2016), Anodonta euscaphys KP187851, Anodonta lucida KF667529 (Song et al. Citation2016), Sinanodonta woodiana HQ283346 (Soroka Citation2010), Arconaia lanceolata KJ144818 (Wang et al. Citation2016a), Cristaria plicata GU944476 (Lee et al. Citation2012), Cuneopsis pisciculus KP273584, Lamprotula coreana JX050180, Lamprotula gottschei KJ018924 (He et al. Citation2016), Lamprotula tortuosa KC109779 (Wang et al. Citation2013), Lanceolaria grayana KJ495725, Lasmigona compressa HM856638 (Breton et al. Citation2011), Lepidodesma languilati KT381195 (Zhou et al. Citation2016), Pyganodon grandis FJ809754 (Breton et al. Citation2009), Unio crassus KY290446 (Burzyński et al. Citation2017), Unio delphinus KT326917 (Fonseca et al. Citation2016), Nodularia douglasiae KM657954 (Wang et al. Citation2016b), Unio pictorum HM014130 (Soroka and Burzyński Citation2010), Unio tumidus KY021078 (Soroka and Burzyński Citation2017), Utterbackia imbecillis HM856637 (Breton et al. Citation2011), and Utterbackia peninsularis HM856636 (Breton et al. Citation2011). Bayesian Inference, as implemented in BEAST (Bouckaert et al. Citation2014) was used to reconstruct the phylogeny. All the records were downloaded, reoriented to the common origin and aligned using ClustalW (Larkin et al. Citation2007). Since these genomes have the same structure and similar gene lengths, the only alignment ambiguities concerned the unassigned regions. However, these were inconsistent and have no influence on the final phylogeny due to complete elimination of columns with missing data. The optimal model of sequence evolution (GTR + G with relaxed, lognormal clock), matching the observed pattern of substitutions was selected, as previously described (Burzyński et al. Citation2017). The MCMC chains were run in quadruplicates for 20 × 106 generations to reach ESS of at least 300 for each parameter. The four runs were convergent so the final tree samples were combined using logcombiner. The Maximum Clade Credibility tree was generated using treeannotator. The tree was visualized in FigTree (Rambaut Citation2009), and the root of the tree was scaled to match that of the recently published mitogenomic analysis (Burzyński et al. Citation2017). All nodes have posterior probabilities of 1.0, except for the ones indicated. The node bars represent 95% CI on node heights.
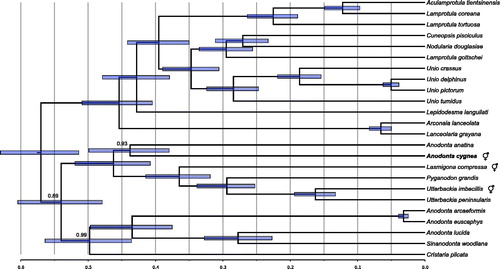
Of the three cases of secondary hermaphroditism covered by the presented data set, the A. cygnea case seems to be the only one without the HORF and also the oldest one (; Mitchell et al. Citation2016). It can be concluded that after the loss of DUI, the supranumerary ORFs can eventually degenerate. This reinforces the hypothesis of the involvement of gender specific mitochondrial ORFs in sex determination of these animals (Breton et al. Citation2011).
Acknowledgements
The authors thank Małgorzata Ożgo (Kazimierz Wielki University in Bydgoszcz, Poland) for help with sample collection.
Disclosure statement
The authors declare no competing interests and are solely responsible for writing this manuscript.
Additional information
Funding
References
- An C, Ouyang S, Zhou C-H, Wu X-P. 2016. The complete F-type mitochondrial genome of Chinese Anodonta arcaeformis (Bivalvia: Unionidae: Anodontinae). Mitochondrial DNA Part A. 27:1552–1553.
- Bogan AE, Roe KJ. 2008. Freshwater bivalve (Unioniformes) diversity, systematics, and evolution: status and future directions. J N Am Benthol Soc. 27:349–369.
- Bouckaert R, Heled J, Kühnert D, Vaughan T, Wu C-H, Xie D, Suchard MA, Rambaut A, Drummond AJ, Prlic A. 2014. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput Biol. 10:e1003537.
- Breton S, Beaupré HD, Stewart DT, Piontkivska H, Karmakar M, Bogan AE, Blier PU, Hoeh WR. 2009. Comparative mitochondrial genomics of freshwater mussels (Bivalvia: Unionoida) with doubly uniparental inheritance of mtDNA: gender-specific open reading frames and putative origins of replication. Genetics. 183:1575–1589.
- Breton S, Stewart DT, Shepardson S, Trdan RJ, Bogan AE, Chapman EG, Ruminas AJ, Piontkivska H, Hoeh WR. 2011. Novel protein genes in animal mtDNA: a new sex determination system in freshwater mussels (Bivalvia: Unionoida)? Mol Biol Evol. 28:1645–1659.
- Burzyński A, Soroka M, Mioduchowska M, Kaczmarczyk A, Sell J. 2017. The complete maternal and paternal mitochondrial genomes of Unio crassus: mitochondrial molecular clock and the overconfidence of molecular dating. Mol Phylogenet Evol. 107:605–608.
- Doucet-Beaupré H, Breton S, Chapman EG, Blier PU, Bogan AE, Stewart DT, Hoeh WR. 2010. Mitochondrial phylogenomics of the Bivalvia (Mollusca): searching for the origin and mitogenomic correlates of doubly uniparental inheritance of mtDNA. BMC Evol Biol. 10:50.
- Fonseca MM, Lopes-Lima M, Eackles MS, King TL, Froufe E. 2016. The female and male mitochondrial genomes of Unio delphinus and the phylogeny of freshwater mussels (Bivalvia: Unionida). Mitochondrial DNA Part B. 1:954–957.
- He F, Wang G. l, Li J. 2016. Complete F-type mitochondrial genome of Chinese freshwater mussels Lamprotula gottschei. Mitochondrial DNA Part A. 27:246–247.
- Hoeh WR, Stewart DT, Guttman SI. 2002. High fidelity of mitochondrial genome transmission under the doubly uniparental mode of inheritance in freshwater mussels (Bivalvia: Unionoidea). Evol Int J Org Evol. 56:2252–2261.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics. 23:2947.
- Lee JH, Choi EH, Kim SK, Ryu SH, Hwang UW. 2012. Mitochondrial genome of the cockscomb pearl mussel Cristaria plicata (Bivalvia, Unionoida, Unionidae). Mitochondrial DNA. 23:39–41.
- Lopes-Lima M, Fonseca MM, Aldridge DC, Bogan AE, Gan HM, Ghamizi M, Sousa R, Teixeira A, Varandas S, Zanatta D, et al. 2017. The first Margaritiferidae male (M-type) mitogenome: mitochondrial gene order as a potential character for determining higher-order phylogeny within Unionida (Bivalvia). J Molluscan Stud. 83:249–252.
- Mitchell A, Guerra D, Stewart D, Breton S. 2016. In silico analyses of mitochondrial ORFans in freshwater mussels (Bivalvia: Unionoida) provide a framework for future studies of their origin and function. BMC Genomics. 17:579.
- Piechocki A, Dyduch-Falniowska A. 1993. Mięczaki (Mollusca). Małże (Bivalvia). [Molluscs (Molusca). Bivalves (Bivalvia) Fauna Słodkowodna Polski. Warszawa (Poland): PWN. Polish.
- Rambaut A. 2009. FigTree, a graphical viewer of phylogenetic trees [Internet]. Institute of Evolutionary Biology, University of Edinburgh. Available from: http://tree.bio.ed.ac.uk/software/figtree
- Skibinski DOF, Gallagher C, Beynon CM. 1994. Mitochondrial DNA inheritance. Nature. 368:817–818.
- Song X-L, Ouyang S, Zhou C-H, Wu X-P. 2016. Complete maternal mitochondrial genome of freshwater mussel Anodonta lucida (Bivalvia: Unionidae: Anodontinae). Mitochondrial DNA Part A. 27:549–550.
- Soroka M. 2010. Characteristics of mitochondrial DNA of unionid bivalves (Mollusca: Bivalvia: Unionidae). I. Detection and characteristics of doubly uniparental inheritance (DUI) of unionid mitochondrial DNA. Folia Malacol. 18:147–188.
- Soroka M, Burzyński A. 2010. Complete sequences of maternally inherited mitochondrial genomes in mussels Unio pictorum (Bivalvia, Unionidae). J Appl Genet. 51:469–476.
- Soroka M, Burzyński A. 2015. Complete female mitochondrial genome of Anodonta anatina (Mollusca: Unionidae): confirmation of a novel protein-coding gene (F ORF). Mitochondrial DNA. 26:267–269.
- Soroka M, Burzyński A. 2017. Doubly uniparental inheritance and highly divergent mitochondrial genomes of the freshwater mussel Unio tumidus (Bivalvia: Unionidae). Hydrobiologia, In press. Available from: https://link.springer.com/article/10.1007/s10750-017-3113-7
- Staden R, Judge DP, Bonfield JK. 2001. Sequence assembly and finishing methods. In: Baxevanis AD, Ouellette BFF, editors. Bioinformatics: a practical guide to the analysis of genes and proteins, Vol. 43, 2nd ed. New York (UK): Wiley.
- Wang G, Cao X, Li J. 2013. Complete F-type mitochondrial genome of Chinese freshwater mussel Lamprotula tortuosa. Mitochondrial DNA. 24:513–515.
- Wang G, Guo L, Li J. 2016a. The F-type complete mitochondrial genome of Arconaia lanceolata. Mitochondrial DNA Part A. 27:322–323.
- Wang G, Xue T, Chen M, Guo L, Li J. 2016b. Complete F-type mitochondrial genome of freshwater mussels Unio douglasiae. Mitochondrial DNA Part A. 27:4021–4022.
- Wu R-W, An C-T, Wu X-P, Zhou C-H, Ouyang S. 2016. Complete maternal mitochondrial genome of freshwater mussel Aculamprotula tientsinensis (Bivalvia: Unionidae: Unioninae). Mitochondrial DNA Part A. 27:4520–4521.
- Zbawicka M, Burzyński A, Wenne R. 2007. Complete sequences of mitochondrial genomes from the Baltic mussel Mytilus trossulus. Gene. 406:191–198.
- Zhou C-H, Ouyang S, Wu X-P, Ding M-H. 2016. The complete maternal mitochondrial genome of rare Chinese freshwater mussel Lepidodesma languilati (Bivalvia: Unionidae: Unioninae). Mitochondrial DNA Part A. 27:4615–4616.
- Zouros E, Ball AO, Saavedra C, Freeman KR. 1994. Mitochondrial DNA inheritance. Nature. 368:818.
