Abstract
Lilium martagon var. pilosiusculum is an endangered species with high ornamental value in Xinjiang province (China). In this study, we reported a complete chloroplast genome of L. martagon var. pilosiusculum, which was de novo assembled using the next-generation sequencing data. The complete chloroplast genome is 152,816 in length, including a large single copy region of 82,265 bp and a small single copy region of 17,541 bp and two inverted repeat regions of 26,505 bp. A total of 110 functional genes were encoded, consisting of 76 protein-coding genes, 30 transfer RNA genes, and four ribosomal RNA genes. The overall AT content of the chloroplast genome is 63.00%. In addition, 68 SSRs and 31 large repeat sequences were found. In the maximum likelihood tree, a strong phylogenetic signal showed that L. martagon var. pilosiusculum is a species of lilium.
Lilium martagon Linnaeus var. pilosiusculum Freyn is a perennial flower bulbs belonging to the section Martagon, the genus Lilium (Liliaceae). L. martagon var. pilosiusculum is characterized by leaves whorled, flowers 2–7 in a raceme, nodding to horizontal, tepals purple-red with deeply coloured spots, nectaries papillose on both surfaces (Liang and Tamura Citation2000). It is a popular ornamental. In China, this species is only distributed in Xinjiang province. However, it is considered as a rare and endangered species in Information System of Chinese Rare and Endangered Plants (ISCREP) (http://rep.iplant.cn/prot/Lilium%20martagon%20var.%20pilosiusculum) and China Biodiversity Red List: Higher Plants (http://www.zhb.gov.cn/gkml/hbb/bgg/201309/t20130912_260061.htm) due to the human activities, unreasonable deforestation and habitat deterioration. Based on the complete sequencing of chloroplast genome, the genetic background can be better understood, which is helpful in promoting the preservation of the species.
A wild individual of L. martagon var. pilosiusculum was collected from Xinjiang province, China. Voucher specimen was deposited in Beijing Agro-Biotechnology Research Center (Beijing, China). Total genomic DNA was extracted from fresh leaves, according to the DNAsecure Plant Kit (Aidlab). A genomic DNA library was constructed using VAHTSTM Turbo DNA Library Prep Kit for Illumina® (Vazyme, Nanjing City, China). High-throughput sequencing was performed with pair-end reads on the HiSeq4000 Sequencing System at Novogene (http://www.novogene.com/index.php). The raw reads were quality-trimmed by NGSQC Toolkit v2.3.3 (Patel and Jain Citation2012) and assembled by SPAdes v3.6.1 (Bankevich et al. Citation2012). Assembled chloroplast genome was annotated using Dual Organellar GenoMe Annotator (http://dogma.ccbb.utexas.edu/) (Wyman et al. Citation2004). The gene map of the cp genome was drawn in OGDraw v1.2 (Lohse et al. Citation2013).
Whole chloroplast genome sequence of L. martagon var. pilosiusculum has been submitted to GenBank with the accession number MF964219. It is 152,816 in length, including a large single copy (LSC) region of 82,265 bp and a small single copy (SSC) region of 17,541 bp and two inverted repeat (IR) regions of 26,505 bp. The complete cp-DNA encodes 110 genes, comprising 76 protein-coding genes, 30 transfer RNA genes, and four ribosomal RNA genes. Among these genes, 15 genes (trnA-UGC, trnG-GCC, trnI-GAU, trnK-UUU, trnL-UAA, trnV-UAC, rps16, petD, atpF, rpl16, petB, rpl2, ndhA, ndhB, and rpoC1) contained one intron, two genes (ycf3 and clpP) contained two introns and 19 genes (rrn4.5, rrn5, rrn16, rrn23, trnA-UGC, trnH-GUG, trnI-CAU, trnI-GAU, trnL-CAA, trnN-GUU, trnR-ACG, trnV-GAC, ndhB, rps7, rpl2, rpl23, ycf2, ycf1, and rps19) were located in IR region. The nucleotide composition of L. martagon var. pilosiusculum has high A + T content of 63.00%, and the corresponding values of the SSC, LSC, and IR regions were 69.30%, 65.20%, and 57.60%, respectively.
The perl program MISA (http://pgrc.ipk-gatersleben.de/misa/) was used to identify the simple sequence repeats (SSRs) loci. The repeat thresholds for mono-, di-, tri-, tetra-, penta-, and hexa-nucleotide SSRs were set to a minimum of 10, 5, 4, 3, 3, and 3 repeats, respectively. The online program REPuter (Kurtz et al. Citation2001) was used to locate the large repeat sequences with a minimal repeat size of 30 bp and hamming distance equal to 3. In this study, 68 SSRs was found, including 38 mono-nucleotide SSRs, 16 di-nucleotide SSRs, four tri-nucleotide SSRs, 10 tetra-nucleotide SSRs. There were no penta- and hexa-nucleotide repeats. In addition, 31 large repeat sequences were identified, containing 11 forward repeats, 18 palindromic repeats, and two reverse repeats. These repeats ranged from 30 to 50 bp in length. SSRs and large repeat sequences can provide important information for research on population genetics and genetic markers.
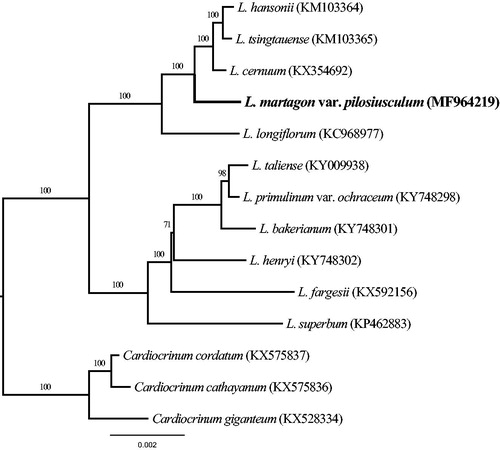
In order to ascertain phylogenetic position of L. martagon var. pilosiusculum, a maximum likelihood (ML) phylogenetic tree was constructed with CIPRES (http://www.phylo.org/) (Miller et al. Citation2010). The complete chloroplast genome of 14 representative species from genera Lilium and Cardiocrinum (the genus Cardiocrinum species as outgroup) were selected to perform the ML analysis () (Kim and Kim Citation2013; Kim et al. Citation2015; Mennes et al. Citation2015; Bi et al. Citation2016; Du et al. Citation2016; Ji et al. Citation2016; Lu et al. Citation2016; Du et al. Citation2017; Zhang et al. Citation2017). The result indicated that 10 of 12 nodes were supported by bootstrap values 100% and the other two nodes by values >71%. In this ML tree, L. martagon var. pilosiusculum is recognized a species of lilium with a strong phylogenetic signal. Moreover, the species is closely related to L. hansonii, L. tsingtauense, L. cernuum, and L. longiflorum in this tree topology structure. The complete chloroplast genome can be subsequently utilized for genetic diversity, identifying species, taxonomy and phylogenetic evolution studies for this species. The available genome information also provides valuable insight into conservation and exploitation efforts for this endangered species.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19:455–477.
- Bi Y, Du YP, Chen XQ, Yang FP, Xue J, Zhang XH, Dong R. 2016. The complete chloroplast genome sequence of Lilium fargesii (Lilium, Liliaceae). Conserv Genet Resour. 8:419–422.
- Du Y, Bi Y, Chen X, Yang F, Xue J, Zhang X. 2016. The complete chloroplast genome of Lilium cernuum: genome structure and evolution. Conserv Genet Resour. 8:375–378.
- Du YP, Bi Y, Yang FP, Zhang MF, Chen XQ, Xue J, Zhang XH. 2017. Complete chloroplast genome sequences of Lilium: insights into evolutionary dynamics and phylogenetic analyses. Sci Rep. 7:5751.
- Ji HS, Chang YY, Do HDK, Lee WB, Kim JH. 2016. The complete chloroplast genome sequence of Lilium tsingtauense Gilg (sect. Martagon, Liliaceae). Mitochondrial DNA B Resour. 1:318–320.
- Kim JS, Kim JH. 2013. Comparative genome analysis and phylogenetic relationship of order Liliales insight from the complete plastid genome sequences of two Lilies (Lilium longiflorum and Alstroemeria aurea). PLoS One. 8:e68180.
- Kim K, Hwang YJ, Lee SC, Yang TJ, Lim KB. 2015. The complete chloroplast genome sequence of Lilium hansonii Leichtlin ex D.D.T.Moore. Mitochondrial DNA. 27:3678–3679.
- Kurtz S, Choudhuri JV, Ohlebusch E, Schleiermacher C, Stoye J, Giegerich R. 2001. REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 29:4633–4642.
- Liang SY, Tamura MN. 2000. Lilium Linnaeus, vol. 24. Beijing (China)/St. Louis (MO): Science Press/Missouri Botanical Garden Press/Flora of China.
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. OrganellarGenomeDRAW—a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41:W575–W581.
- Lu RS, Li P, Qiu YX. 2016. The complete chloroplast genomes of three Cardiocrinum (Liliaceae) species: comparative genomic and phylogenetic analyses. Front Plant Sci. 7:2054.
- Mennes CB, Lam VKY, Rudall PJ, Lyon SP, Graham SW, Smets EF, Merckx VSFT. 2015. Ancient Gondwana break-up explains the distribution of the mycoheterotrophic family Corsiaceae (Liliales). J Biogeogr. 42:1123–1136.
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Paper presented at: the Gateway Computing Environments Workshop (GCE); Nov 14.
- Patel RK, Jain M. 2012. NGS QC toolkit: a toolkit for quality control of next generation sequencing data. PLoS One. 7:e30619.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Zhang Q, Bi Y, Zhang MF, Chen XQ, Yang FP, Xue J, Du YP, Zhang XH. 2017. The complete chloroplast genome of Lilium taliense, an endangered species endemic to China. Conserv Genet Resour. 9:201–203.