Abstract
Psathyrostachys huashanica Keng belongs to the genus Psathyrostachys within the Gramineae family, is a rare and endangered herb plant endemic to China. In the current work, we first developed its whole chloroplast (cp) genome sequence. The cp genome of P. huashanica is 136,632 bp in size, including a large single copy (LSC) region of 81,803 bp, a small single copy (SSC) region of 12,763 bp, and a pair of inverted repeats (IRs) of 21,033 bp. The genome contains 114 genes, including 78 protein-coding genes, 32 tRNA genes, and 4 rRNA genes. The GC content in cp genome, LSC, SSC, and IR regions were 38.5, 36.5, 32.6, and 38.5%, respectively. The phylogenetic analysis showed P. huashanica is closely related to Triticum genus.
Psathyrostachys huashanica Keng belongs to the Psathyrostachys genus within the Gramineae family, which is a rare and endangered herb plant endemic to Huashan Mountain, Shaanxi, China (Hang et al. Citation2004). As a wildly relative species of wheat, P. huashanica has the crucial values. This herb species has also important cold and drought resistances (Du Citation2010; Li et al. Citation2011). However, in recent years, due to the human activities and rapidly climate oscillation, results in the increasingly declining of P. huashanica resources. It is thus urgent to take conservation and management strategies to conserve this endangered herb plant. Herein, we first characterized the complete chloroplast (cp) genome of P. huashanica.
Fresh plant leaves of P. huashanica were sampled from Huashan Mountain, Shaanxi, China (N34.58°, E110.09°), and the materials and voucher specimen (No. PHLZH2014113) were stored in Northwest University. Total DNA was extracted with improved CTAB method (Li et al. Citation2009), and then used for the next-generation sequencing with Illumina Hiseq 2500 platform by Sangon Biotech (Shanghai, China). All the raw reads were trimmed using the program NGSQCToolkit_version 2.3.3 (Patel and Jain Citation2012). After dislodged the low quality reads, the clean reads were assembled by MIRA version 4.0.2 (Chevreux et al. Citation2004), and MITObim version 1.8 (Hahn et al. Citation2013) with the cp genome of closely related species Hordeum vulgare (EF115541) as the reference. A total of 117,623 individual reads have been assembled, and the average coverage was 131.8×. The complete cp genome was annotated using the online software DOGMA and then compared with the cp genome of H. vulgare (EF115541) to manually correct in Geneious version 8.0.2 (Kearse et al. Citation2012). Eventually, the whole genome sequence of P. huashanica has been submitted to GenBank with the accession number MK135434.
The cp genome of P. huashanica is 136,632 bp in length, including a large single copy (LSC) region of 81,803 bp, a small single copy (SSC) region of 12,763 bp and a pair of inverted repeats (IRs) of 21,033 bp (). The GC content in cp genome, LSC region, SSC region, and IR region are 38.5, 36.5, 32.6, and 38.5%, respectively, and it is similar to other Gramineae plants. The cp genome contains 114 genes, including 78 protein-coding genes, 32 tRNA genes, and 4 r RNA genes. But the genes accD, ycf1, and ycf2 are degraded. A total of 15 genes (trnA-UGC, trnG-UCC, trnI-GAU, trnK-UUU, trnL-UAA, trnV-UAC, rps12, rps16, rpl2, rpl16, ndhA, ndhB, petB, petD, and atpF) contain a single intron, and only one gene (ycf3) contains two introns.
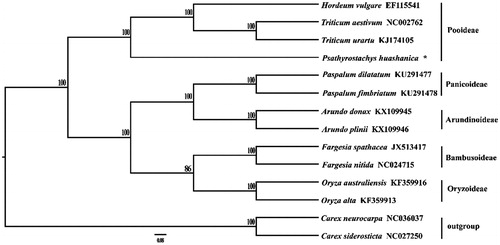
A total of 14 available cp genomes were used to construct the phylogenetic tree among the most of Gramineae species with Carex siderosticta (NC_027250) and Carex neurocarpa (NC_036037) as outgroups (). The programme Modeltest version 3.7 (Posada and Crandall Citation1998) was used to determine the best-fitting model based on the Akaike information criterion. Maximum likelihood analysis was performed using RAxML version 7.2.8 (Stamatakis Citation2006) with 500 bootstrap replicates. The results showed that P. huashanica is closely related to the genus Triticum.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Chevreux B, Pfisterer T, Drescher B, Driesel AJ, Müller WE, Wetter T, Suhai S. 2004. Using the miraEST assembler for reliable and automated mRNA transcript assembly and SNP detection in sequenced ESTs. Genome Res. 14:1147–1159.
- Du YN. 2010. Drought-resistance preliminary research and cytological identification on the progeny of Triticum-Psathyrostachys. Shaanxi, China: Northwest A&F University.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads-a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129.
- Hang Y, Jin Y, Lu BR. 2004. Genetic diversity of the endangered species Psathyrostachys huashanica in China and its strategic conservation. J Fudan Univ. 43:260–266.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Li Q, Liu X, Yue M, Meng QC. 2011. Seed germination and seedling physiological characteristics in Psathyrostachys huashanica Keng under the drought and salt stress. Acta Bot Boreal-Occident Sin. 31:319–324.
- Li RH, Xia YS, Liu SZ, Sun LL, Guo PG, Miao SY, Chen JH. 2009. CTAB improved method of DNA extraction in plant. Res Exploration Lab. 28:14–16.
- Patel RK, Jain M. 2012. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS One. 7:e30619.
- Posada D, Crandall KA. 1998. Modeltest: testing the model of DNA substitution. Bioinformatics. 14:817–818.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22:2688–2690.