Abstract
The fish family Scombropidae consists of a single genus, Scombrops, and is distributed in the waters of the northwestern Pacific Ocean, southwestern Indian Ocean and western Atlantic Ocean, including the Caribbean Sea. Among these, the population of South Africa has been renamed from Scombrops dubius to Scombrops boops, which is known to be distributed in waters around Japanese Archipelago. We are skeptical of this revision as the two populations are geographically isolated between the waters of the Far East and the southern end of the Africa coast. Recently, our lab determined the sequences of the complete mitochondrial genome of three Japanese gnomefish, S. boops, Scombrops gilberti and an undescribed scombropid species (Scombrops sp.). Here, we compared the partial sequence of cytochrome c oxidase subunit I (COI) from the three Japanese gnomefish with that of the African population. The African sequences showed 95.5–96.6% identity with the Japanese sequences, while the corresponding sequences from the Japanese species showed 98.2–100% identity with each other. A maximum likelihood tree based on the partial sequence of COI also demonstrated that the clade of African gnomefish is distinct from those of Japanese species including S. boops. These results suggest that the scombropid fish in the African waters is a different species from S. boops and that it is appropriate to resurrect the name Scombrops dubius for the African population.
Introduction
The taxonomic classification of fishes has traditionally been based on morphological and meristic characteristics. However, advances in molecular biology have made possible rapid developments in molecular phylogenetics and associated tools. For example, among the more recent tools is the DNA barcoding of eukaryotes by targeting the conserved region of the cytochrome c oxidase subunit I (COI) gene in the mitochondrial DNA (mtDNA), a concept that was introduced about 15 years ago (Hebert et al. Citation2003a). Thereafter, copious amounts of related data have been accumulating rapidly in DNA databases, and many studies have been published using this technology (Hebert et al. Citation2003b; Ward et al. Citation2008; Steinke et al. Citation2009, Citation2016; Ma et al. Citation2012; Zhu et al. Citation2013; Khedkar et al. Citation2014; Chen et al. Citation2015; Dhar and Ghosh Citation2015). Taxonomic resolution and accuracy of species identification have improved greatly with the accumulation of barcode-associated sequence data (Zhang and Hanner Citation2012). The availability of DNA barcoding and related tools should contribute to finding cryptic species and a better understanding of their ecology (Zhang and Hanner Citation2012; Kress et al. Citation2015).
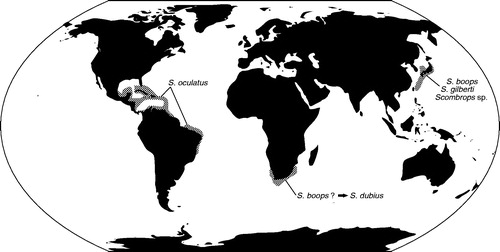
Recently, our laboratory reported a cryptic gnomefish species collected from the southern waters off the Japanese Archipelago, based on the mitochondrial cytochrome b gene sequence (Itoi et al. Citation2018). This finding revealed, for the first time, that three species of the genus Scombrops are distributed in the narrow waters of the Far East, suggesting that the classification of the genus Scombrops worldwide is questionable. Gnomefish belong to the genus Scombrops and have been thought to include four species worldwide, distributed in the waters of the northwestern Pacific Ocean, southwestern Indian Ocean and western Atlantic Ocean including the Caribbean Sea (Poey Citation1860; Yasuda et al. Citation1971; Mochizuki Citation1979; Heemstra, Citation1986; Itoi et al. Citation2008, Citation2018). Furthermore, it has been thought that Scombrops boops, Scombrops gilberti, and Scombrops sp. are distributed in waters around the Japanese Archipelago (Yasuda et al. Citation1971; Itoi et al. Citation2008, Citation2010, Citation2011, Citation2018; Noguchi et al. Citation2012; Takai et al. Citation2014). It has also been shown that the gnomefish off Africa, which was called Scombrops dubius, is actually S. boops (Heemstra Citation1986). However, we are skeptical that the two populations of S. boops, isolated from each other between the waters of northwestern Pacific Ocean and those of southwestern Indian Ocean, and with their distributions restricted to their respective geographic locations, are the same species ().
Figure 1. Geographic distribution of the scombropid fishes. Areas of distribution are shaded in gray.
Therefore, we used the COI gene sequence, generally used for DNA barcoding of eukaryotes as mentioned above, to determine if the African and Japanese populations of S. boops are two distinct species or two geographically separated populations of the same species. Although the complete mtDNA sequence is available for the three Japanese gnomefishes (Tsunashima et al. Citation2016a, Citation2016b; Mochizuki et al. Citation2017), it is not for the African population. The identity of the complete mtDNA sequences is approximately 97% among the three Japanese scombropid species, although the sensitivity for species identification differs among the genes/regions of the sequence (Mochizuki et al. Citation2017). We report the results of phylogenetic analysis based on the sequence of COI gene, which reveals genetic differences among the scombropids.
Materials and methods
Fish sample
A total of 37 specimens of Japanese scombropids were used: three species S. boops (n = 13), S. gilberti (n = 1) and Scombrops sp. (n = 9) were captured at southern waters off Kyushu Island, Japan (Table S1). Additional specimens of S. boops (n = 3) and Scombrops sp. (n = 1) were collected at the waters around Yonagunijima Island, while those of S. gilberti (n = 10) were collected from the waters off Iwate, Japan (Figure S1; Table S1). Fish were stored at −20 °C until dissection.
DNA extraction and PCR amplification
Total genomic DNA was extracted from the muscle of 37 scombropid specimens, using the method of Itoi et al. (Citation2018). Partial fragment of COI gene was amplified by PCR using primers Sc_COI_F1 (5′-CGACTAATCACAAAGACATCGGCAC-3′) and Sc_COI_R1 (5′-AAACCTCTGGGTGACCAAAGAATCA-3′), which were designed based on the mtDNA sequences from three Japanese scombropid species. PCR amplification was performed using a reaction mixture containing genomic DNA as a template, 4 μL of 5 × GoTaq DNA polymerase buffer (Promega), 2.6 μL of 5 mM primers, 2 μL of 2 mM dNTP, 2 μL of 25 mM MgCl2 and one unit of GoTaq DNA polymerase (Promega) brought to a total volume of 20 μL with sterile water. The thermal cycling profile for the PCR consisted of initial denaturation at 94 °C for 3 min followed by 40 cycles of denaturation at 94 °C for 15 s, annealing at 50 °C for 20 s and extension at 72 °C for 45 s.
Sequencing of PCR products and phylogenetic analysis
Sequencing of PCR products was performed for both strands with a 3130xl Genetic Analyzer (Applied Biosystems) using a BigDye Terminator v3.1 Cycle Sequencing Ready Reaction Kit (Applied Biosystems). Visual inspection of DNA sequence chromatograms was carried out using a program, MEGA ver. 7.0.26 (Kumar et al. Citation2016), to check sequence quality and correct base-calling and sequencing errors. Alignment of partial sequences of the COI gene for scombropid fishes obtained in this study was performed using Clustal Omega (Sievers et al. Citation2011). The sequences of the COI gene have been submitted to the DDBJ/EMBL/GenBank databases under the accession numbers LC388035–LC388071 and the trace files were deposited in BOLD system under the process IDs JPG004-19–JPG040-19. Maximum likelihood tree was constructed using MEGA ver. 7.0.26 for the data on the nucleotide sequences of the scombropid COI gene (652 bp).
Results and discussion
The partial sequence of COI gene was amplified by PCR from 16 specimens of S. boops, 11 specimens of S. gilberti, and 10 specimens of the undescribed gnomefish from the Japanese waters. A BLAST search of the nucleotide sequences of the COI for the three gnomefish species showed near identity (98.2–100%) to the corresponding sequences in DDBJ/EMBL/GenBank (). The COI sequences from three Japanese species were then compared with those from African populations of S. boops available in the database (JF494461, HQ945916). The COI sequences from African population were 95.6–96.2, 96.3–96.6, and 95.6–96.0% identical to those from S. boops, S. gilberti, and the undescribed species, respectively (). The phylogenetic tree indicates that the African clade is distinct from the Japanese species (), suggesting genetic differences between the two scombropid groups. A previous study from our laboratory compared the complete mtDNA sequence among the three Japanese gnomefish species and reported that the percent identity among them was 94.4–99.9% for protein-coding genes, 90.5–100% for tRNA genes, 99.2–99.5% for rRNA genes, and 92.8–94.9% for control region (Mochizuki et al. Citation2017), suggesting that the resolution for the classification of gnomefish species depends on the gene/region used in the analysis. Recently, Steinke et al. (Citation2016) were able to successfully identify South African marine fishes from various stages of life, including eggs, larvae, and adults, based on nucleotide sequences of COI linked to authoritative voucher specimens. Therefore, we presume that COI sequences are appropriate for species identification in the genus Scombrops as well.
Figure 2. Phylogenetic relationship of the Japanese scombropid species and African population inferred from the sequence of cytochrome c oxidase subunit I (COI) gene. The phylogenetic tree was generated by maximum likelihood analysis under the General Time Reversible model. Numbers above the branches denote the bootstrap percentages from 1000 replicates. The accession numbers for the sequences are shown in parentheses. The accession number LC388035–LC388071 refer to those deposited in the DDBJ/EMBL/GenBank. Epigonus pandionis (KT883637), Epigonus denticulatus (AP017435, JF493426) and Epigonus telescopus (KJ709756) were used as the outgroup in the analysis. Only bootstrap probabilities >50% are shown. The scale at the bottom of the tree shows the number of nucleotide substitutions per site.
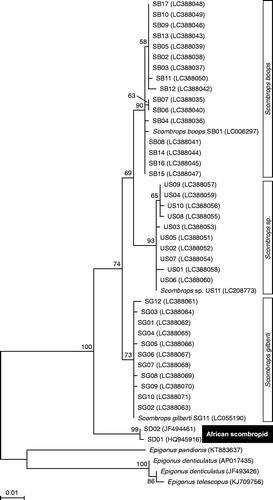
Table 1. Identities between the nucleotide sequence of COI gene (652 bp) from species-identified sample and those from gnomefishes on DNA databases.
The classification of the genus Scombrops even in the Japanese populations has been ambiguous because of the similarities in morphology (Jordan and Snyder Citation1901a, 1901Citationb; Tanaka Citation1931; Oshima Citation1939). Although the genus Scombrops was not accompanied by a species name in the original description, the species name Scombrops cheilodipteroides was first mentioned by Bleeker (Citation1854). However, S. boops became the established name perhaps because S. cheilodipteroides and Sparus boops referred to the same species (Jordan and Snyder Citation1901a). Subsequently, Telescopias gilberti was distinguished from the S. boops based on the difference in their external characters (Jordan and Snyder Citation1901b). On the other hand, Telescopias gilberti was used as a synonym of S. boops by Tanaka (Citation1931) and followed subsequently by several authors. Later, Yasuda et al. (Citation1971) reported that the meristic and morphometric characters were different between S. boops and T. gilberti, and the name S. gilberti became established. These differences were subsequently supported by genetic differences based on mtDNA sequences (Itoi et al. Citation2008).
Recently, an undescribed species was detected by our laboratory from the Japanese scombropid populations that include S. boops and S. gilberti, based on the difference in the relationship between otolith weight and standard length as well as the nucleotide sequences of cytochrome b gene (Itoi et al. Citation2018). The complete mtDNA sequences of the undescribed species were 97% and 96% identical to S. boops and S. gilberti, respectively (Mochizuki et al. Citation2017). Interestingly, the African scombropid population called Scombrops dubius has been considered the synonym of S. boops (Heemstra Citation1986). However, it is difficult to imagine that the same species is distributed in the waters around the Japanese Archipelago and off South Africa/Mozambique. This study demonstrates that the partial sequences of COI of the Japanese scombropid species are more related to each other than to those of the African scombropid, suggesting that the African scombropid is a different from the Japanese species. Therefore, we propose that the species name Scombrops dubius would be resurrected for the African population. Further investigation would be necessary to unravel the classification of these scombropid species.
Furthermore, the detection of the undescribed species from the Japanese scombropids suggests that the number of species belonging to the genus Scombrops worldwide might be an underestimate. It is generally known that there is rich species diversity of marine organisms around the northwestern Pacific Ocean (Fujikura et al. Citation2010). It is likely that the populations in western Atlantic Ocean and in the waters off South Africa/Mozambique might also include some cryptic species as a result of the extremely wide range of their distribution.
Supplemental Material
Download Zip (330.6 KB)Disclosure statement
The authors declare that there are no conflicts of interest.
Additional information
Funding
References
- Bleeker P. 1854. Nieuwe nalezingen op de ichthyologie van Japan. Verh Bat Gen. 26:1–132. pls. 1–8.
- Chen W, Ma X, Shen Y, Mao Y, He S. 2015. The fish diversity in the upper reaches of the Salween River, Nujiang River, revealed by DNA Barcoding. Sci Rep. 5:17437.
- Dhar B, Ghosh SK. 2015. Genetic assessment of ornamental fish species from North East India. Gene. 555:382–392.
- Fujikura K, Lindsay D, Kitazato H, Nishida S, Shirayama Y. 2010. Marine biodiversity in Japanese waters. PLoS One. 5:e11836.
- Hebert PDN, Ratnasingham S, deWard JR. 2003a. Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proc R Soc Lond B. 270:S96–S99.
- Hebert PDN, Cywinska A, Ball SL, deWaard JR. 2003b. Biological identifications through DNA barcodes. Proc Biol Sci. 270:313–321.
- Heemstra PC. 1986. Scombropidae. In: Smith MM, Heemstra PC, editors. Smiths’ sea fishes. Berlin: Springer-Verlag; p. 563.
- Itoi S, Mochizuki Y, Tanaka M, Oyama H, Tsunashima T, Yamada R, Shishido H, Masuda Y, Nakai S, Takai N, et al. 2018. Species composition of the genus Scombrops (Teleostei, Scombropidae) in the waters around the Japanese Archipelago: detection of a cryptic species. Mitochondrial DNA Part A. 29:1293–1300.
- Itoi S, Odaka J, Noguchi S, Noda T, Yuasa K, Muraki T, Tanabe T, Takai N, Yoshihara K, Sugita H. 2011. Genetic homogeneity between adult and juvenile populations of Scombrops gilberti (Percoid, Scombropidae) in the Pacific Ocean off the Japanese Islands. Fish Sci. 77:975–981.
- Itoi S, Odaka J, Yuasa K, Akeno S, Nakajima A, Suenaga A, Noda T, Akimoto S, Myojin T, Ikeda Y, et al. 2010. Distribution and species composition of juvenile and adult scombropids (Teleostei, Scombropidae) in Japanese coastal waters. J Fish Biol. 76:369–378.
- Itoi S, Takai N, Naya S, Dairiki K, Yamada A, Akimoto S, Yoshihara K, Sugita H. 2008. A species identification method for Scombrops boops and Scombrops gilberti based on polymerase chain reaction-restriction fragment length polymorphism analysis of mitochondrial DNA. Fish Sci. 74:503–510.
- Jordan DS, Snyder JD. 1901a. A list of Japanese fishes. Proc U S Nat Mus. 23:739–769. pls 31–38.
- Jordan DS, Snyder JD. 1901b. Cardinal fishes of Japan. Proc U S Nat Mus. 24:891–913. pls 63–64.
- Khedkar GD, Jamdade R, Naik S, David L, Haymer D. 2014. DNA barcodes for the fishes of the Narmada, one of India's longest rivers. PLoS One. 9:e101460.
- Kress WJ, García-Robledo C, Uriarte M, Erickson DL. 2015. DNA barcodes for ecology, evolution, and conservation. Trends Ecol. Evol. (Amst.). 30:25–35.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Ma H, Ma C, Ma L. 2012. Molecular identification of genus Scylla (Decapoda: Portunidae) based on DNA barcoding and polymerase chain reaction. Biochem Syst Ecol. 41:41–47.
- Mochizuki K. 1979. Age and growth of the two Japanese scombropids, Scombrops boops and S. gilberti. Jpn J Ichthyol. 26:62–68.
- Mochizuki Y, Yamada R, Shishido H, Masuda Y, Nakai S, Takai N, Itoi S, Sugita H. 2017. Complete mitochondrial genome of an undescribed gnomefish of the genus Scombrops (Teleostei, Scombropidae) from southern waters off Kyushu Island, Japan. Mitochondrial DNA Part B. 2:106–108.
- Noguchi S, Itoi S, Takai N, Noda T, Myojin T, Yoshihara K, Sugita H. 2012. Population genetic structure of Scombrops boops (Percoid, Scombropidae) around the Japanese archipelago inferred from the cytochrome b gene sequence in mitochondrial DNA. Mitochondrial DNA. 23:233–239.
- Oshima M. 1939. Fish. Tokyo: Sanseido. 661 pp. (in Japanese).
- Poey F. 1860. Poissons de Cuba. Mem Hist Nat Isla Cuba. 2:115–356.
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, et al. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 7:539.
- Steinke D, Connell AD, Hebert PDN. 2016. Linking adults and immatures of South African marine fishes. Genome. 59:959–967.
- Steinke D, Zemlak TS, Boutillier JA, Hebert PDN. 2009. DNA barcoding of Pacific Canada’s fishes. Mar Biol. 156:2641–2647.
- Takai N, Kozuka Y, Tanabe T, Sagara Y, Ichihashi M, Nakai S, Suzuki M, Mano N, Itoi S, Asahina K, et al. 2014. Habitat use of the gnomefishes Scombrops boops and S. gilberti in the northwestern Pacific Ocean in relation to reproductive strategy. Aquat Biol. 21:109–120.
- Tanaka S. 1931. On the distribution of fishes in Japanese waters. J Fac Sci Imp Univ Tokyo Sec 4 Zool. 3:1–90. pls 1–3.
- Tsunashima T, Itoi S, Abe K, Takigawa T, Inoue S, Kozen T, Ono N, Noguchi S, Nakai S, Takai N, et al. 2016a. The complete mitochondrial genome of the gnomefish Scombrops boops (Teleostei, Perciformes, Scombropidae) from the Pacific Ocean off the Japanese Islands. Mitochondrial DNA Part A. 27:785–786.
- Tsunashima T, Yamada R, Abe K, Noguchi S, Itoi S, Nakai S, Takai N, Sugita H. 2016b. Phylogenetic position of Scombropidae within teleostei: the complete mitochondrial genome of the gnomefish, Scombrops gilberti. Mitochondrial DNA Part A. 27:3446–3448.
- Ward RD, Costa FO, Holmes BH, Steinke D. 2008. DNA barcoding of shared fish species from the North Atlantic and Australasia: minimal divergence for most taxa, but Zeus faber and Lepidopus caudatus each probably constitute two species. Aquat Biol. 3:71–78.
- Yasuda F, Mochizuki K, Kawajiri M, Nose Y. 1971. On the meristic and morphometric differences between Scombrops boops and S. gilberti. Jpn J Ichthyol. 18:118–124.
- Zhang J, Hanner R. 2012. Molecular approach to the identification of fish in the South China Sea. PLoS One. 7:e30621.
- Zhu SR, Fu JJ, Wang W, Li JL. 2013. Identification of Channa species using the partial cytochrome c oxidase subunit I (COI) gene as a DNA barcoding marker. Biochem Syst Ecol. 51:117–123.
