Abstract
In this study, the complete plastid genome of Fragilariopsis cylindrus was recovered through Illumina sequencing data. This complete plastid genome of F. cylindrus was 123,275 bp in length and contained a pair of IR regions (13,063 bp). It encoded 141 genes including 104 protein-coding genes, 30 tRNA genes, one tmRNA, and six ribosomal RNA genes in IR regions. The overall GC content of F. cylindrus pt genome is 30.8%. By phylogenetic analysis, using whole genome alignments through NJ method, F. cylindrus exhibited the closest relationship with Pseudo-nitzschia multiseries.
Fragilariopsis cylindrus is a species of diatom in the family Bacillariaceae. The genome sequence of Fragilariopsis cylindrus was published in 2017 (Thomas et al. Citation2017). However, the organelle genome information of F. cylindrus is still limited.
The sample of Fragilariopsis cylindrus strain CCMP1102 was obtained from the National Centre for Marine Algae and Microbiota and was maintained in lab environment. The isolated DNA was stored in the sequencing company (HuiTong Tech, Shenzhen, China) and sequencing was carried out using Illumina Hiseq 2000 platform.
After reads quality filtration, the clean reads were assembled using SPAdes 3.9.0 (Bankevich et al. Citation2012) based on default settings. To fill the gap, Price (Ruby et al. Citation2013) and MITObim v1.8 (Hahn et al. Citation2013) were applied and Bandage (Wick Ryan et al. Citation2015) was used to identify the borders of the IR regions. The complete sequence was primarily annotated by Plann (Huang et al. Citation2015) combined with manual correction. All tRNAs were confirmed using the tRNAscan-SE search server (Lowe et al. Citation1997). Other protein-coding genes were verified using BLAST search on the NCBI website (http://blast.ncbi.nlm.nih.gov/), and manual correction for start and stop codons were conducted. This complete plastid genome sequence together with gene annotations were submitted to GenBank under the accession numbers of MK217824.
The plastid genome of Fragilariopsis cylindrus, with a length of 123,275 bp, has a quadripartite structure compared with other diatom algae. The whole pt genome contains a two inverted repeat (IRs) regions of 4,604 bp, in which encodes two rRNA operons. The pt genome possesses 141 genes, including 104 protein-coding genes, six ribosomal RNA genes (three rRNA species) and 30 tRNA genes and one tmRNA. The overall GC content of the pt genome is 30.8%.
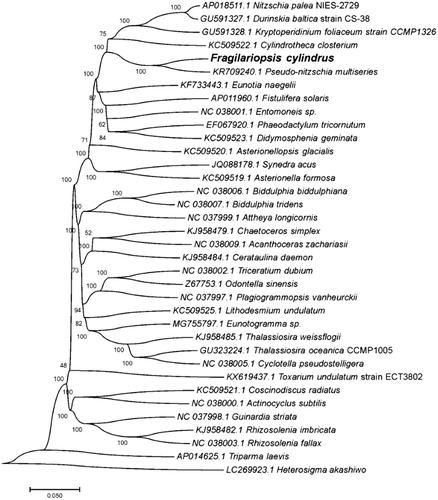
For phylogeny analysis, the genome-wide alignment of 36 algae plastid genomes was carried out using HomBlocks (Guiqi et al. Citation2018) under the Gblocks trimming method, which only conserved phylogenetic informative positions in alignments, resulting in 36,769 positions in total. And, the tree was constructed using NJ method through MEGA6.0 (Koichiro et al. Citation2013) under Maximum Composite Likelihood method with 1000 bootstrap calculations. As expected, F. cylindrus exhibited the closest relationship with Pseudo-nitzschia multiseries ().
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Anton B, Sergey N, Dmitry A, Gurevich AA, Mikhail D, Kulikov AS, Lesin VM, Nikolenko SI, Son P, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19:455–477.
- Bi G, Mao Y, Xing Q, Cao M. 2018. HomBlocks: a multiple-alignment construction pipeline for organelle phylogenomics based on locally collinear block searching. Genomics. 110:18–22.
- Christoph H, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads-a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129–e129.
- Graham RJ, Bellare P, DeRisi JL. 2013. PRICE: software for the targeted assembly of components of (Meta) genomic sequence data. G3 Genes Genomes Genet. 3:865–880.
- Huang DI, Cronk QCB. 2015. Plann: a command-line application for annotating plastome sequences. Appl Plant Sci. 3:1500026.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964.
- Mock T, Otillar RP, Strauss J, McMullan M, Paajanen P, Schmutz J, Salamov A, Sanges R, Toseland A, Ward BJ, et al. 2017. Evolutionary genomics of the cold-adapted diatom Fragilariopsis cylindrus. Nature. 541:536.
- Wick Ryan R, Schultz Mark B, Justin S, Holt KE. 2015. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics. 31:3350–3352.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.