Abstract
The whole chloroplast (cp) genome sequence of Populus nigra L. has been characterized from Illumina pair-end sequencing. The complete cp genome was 156,451 bp in length, containing a large single copy region (LSC) of 84,529 bp, and a small single copy region (SSC) of 16,580 bp, which were separated by a pair of 27,671 bp inverted repeat regions (IRs). The genome contained 144 genes, including 99 protein-coding genes (84 PCG species), 37 tRNA genes (30 tRNA species), and 8 ribosomal RNA genes (4 rRNA species). Most of the gene species occur as a single copy, while 26 gene species occur in double copies. The overall AT content of P. nigra cp genome is 63.2%, while the corresponding values of the LSC, SSC, and IR regions are 65.4, 69.5, and 58.0%, respectively. Further, phylogenetic analysis suggested that P. nigra is closely related to the species of P. alba.
Populus nigra L., the black poplar, is a species of cottonwood poplar, the type species of section Aigeiros of the genus Populus, native to Europe, southwest and central Asia, and northwest Africa (Smulders et al. Citation2008). The P. nigra has the characteristics of a cylindrical crown and towering trunk that looks very beautiful. Another advantage of black poplar is the bark, which can be used as medicine and is very useful for colds, rheumatism, foot emphysema, and burn. Unfortunately, large areas of this natural habitat have been lost due to drainage of rivers, management of riverbanks, intensive grazing, and wood cutting, the wild resources of P. nigra have been dramatically decreased and need urgent conservation (van der Schoot et al. Citation2000). At the same time, the genetics information and the evolutionary relationship of black poplar are still a blank at present. In this study, we assembled and characterized the complete chloroplast genome sequence of P. nigra based on the Illumina pair-end sequencing data.
The voucher specimen of P. nigra was stored at the Institute of Botany, The Chinese Academy of Sciences. The fresh leaves of a single individual of P. nigra were sampled from Altay (Xinjiang, China; 87°49′21.3″E, 47°21′48.3″N), and were used for the total genomic DNA extraction with the modified CTAB method (Doyle Citation1987). The whole-genome sequencing was conducted with 150 bp pair-end reads on the Illumina Hiseq Platform (Illumina, San Diego, CA). In total, about 800 MB high-quality clean reads were obtained and used for the cp genome assembly using Velvet software (Zerbino and Birney Citation2008). The resulting contigs were linked based on overlapping regions after being aligned to P. trichocarpa (EF489041) (Tuskan et al. Citation2006) and visualized in Geneious version 8.0.5 (Kearse et al. Citation2012). Annotation was performed with a newly developed command-line application called Plann (Plastome Annotator), which is suitable for annotation of plastomes with a well-annotated sequenced relative (Huang and Cronk Citation2015). Then, we corrected the annotation with Geneious (Kearse et al. Citation2012). Next, we generate a physical map of the genome using OGDRAW (http://ogdraw.mpimp-golm.mpg.de/) (Lohse et al. Citation2013). A maximum likelihood (ML) tree with 100 bootstrap replicates was inferred using RAxML version 8 (Stamatakis Citation2014) from alignments created by the MAFFT (Katoh and Standley Citation2013) using plastid genomes of 14 species (). The complete chloroplast genome sequence together with gene annotations was submitted to GenBank under the accession numbers of KX377117 for P. nigra.
Figure 1. The phylogenetic tree based on the 14 complete chloroplast genome sequences. Accession numbers: Populus euphratica NC_024747, Populus trichocarpa F489041, Populus fremontii NC_024734, Populus balsamifera NC_024735, Populus cathayana KP729175, Populus tremula KP861984, Populus alba AP008956, Populus yunnanensis KP729176, Salix interior NC_024681, Salix babylonica NC_028350, Salix purpurea NC_026722, Salix suchowensis KM983390, I. polycarpa KX229742.
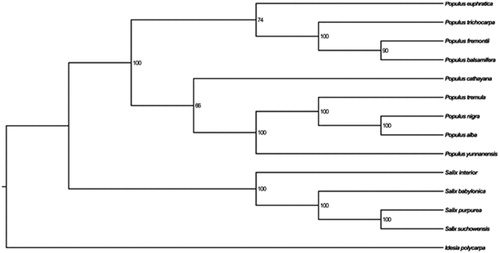
The phylogenetic analysis of 14 plastid genomes which Idesia polycarpa (Yang et al. Citation2016) was used as the outgroup showed that the cp genome of P. nigra is closely related to the species of P. alba (). This complete chloroplast genome can be subsequently used for population, phylogenetic and cp genetic engineering studies of P. nigra, and such information would be fundamental to develop potential new medicinal and ornamental value for this important species.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Doyle JJ. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Huang DI, Cronk QC. 2015. Plann: a command-line application for annotating plastome sequences. Appl Plant Sci. 3:1–3.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. OrganellarGenomeDRAW-a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41:W575–W581.
- Smulders MJM, Cottrell JE, Lefèvre F, Schoot JVD, Arens P, Vosman B, Tabbener HE, Grassi F, Fossati T, Castiglione S. 2008. Structure of the genetic diversity in black poplar (Populus nigra L.) populations across European river systems: consequences for conservation and restoration. For Ecol Manage. 255:0–1399.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Tuskan GA, Difazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam N, Ralph S, Rombauts S, Salamov A, et al. 2006. The genome of black cottonwood, Populus trichocarpa (Torr. &; Gray). Science. 313:1596–1604.
- van der Schoot J, Pospíšková M, Vosman B, Smulders MJM. 2000. Development and characterization of microsatellite markers in black poplar (Populus nigra L.). Theor Appl Genet. 101:317–322.
- Yang W, Wang W, Zhang L, Chen Z, Guo X, Ma T. 2016. Characterization of the complete chloroplast genome of Idesia polycarpa. Conserv Genet Resour. 8:271–273.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829.
