Abstract
Neocaridina heteropoda koreana Kubo, 1938 is a freshwater shrimp native to the Korean peninsula. We have determined the mitogenome of N. heteropoda koreana, which's length is 15,558 bp long including 13 protein-coding genes, 2 ribosomal RNA genes, 22 transfer RNAs, and a single large non-coding region of 674 bp. Its GC ratio is 33.0%. Gene order of N. heteropoda koreana is identical to those of other known Atyidae species. Phylogenetic trees show that N. heteropoda koreana is sister to N. davidi and placed within genus Caridina. Our mitogenome will be a useful resource for understanding molecular phylogeny of genus Neocradina.
The genus Neocaridina are freshwater shrimps native to east Asia, including Korea, China, and Japan (Liang Citation2004). They were traditionally used for fish baits and food ingredients (Crowl et al. Citation2001; Mugo-Bundi et al. Citation2015; Snyder et al. Citation2015). Recently, some species, such as Neocaridina davidi and Neocaridina palmata, have been captive bred as aquarium pets for their brilliant colors and used as biological-control agents for algae in aquariums. In contrast to their active utilizations, the taxonomical position of the genus was not confirmed completely by both morphological and molecular characters (von Rintelen et al. Citation2012). Recent study has even presented cryptic species using mitochondrial genes as markers (Shih et al. Citation2017), therefore requiring additional researches of the taxa; however, mitogenome is available for only one species, N. davidi (formerly N. denticulata sinensis; Yu et al. Citation2014). To fill this shortage, we have assembled the mitochondrial genome of Korean swamp shrimp, Neocaridina heteropoda koreana.
Total DNA of N. heteropoda koreana, collected in Geojedo, Gyeongsangnam province (34°49′00.1″N 128°38′10.0″E), Republic of Korea, was extracted from the abdomen of a single adult shrimp, using DNeasy Blood & Tissue Kit (QIAGEN, Hilden, Germany). Raw sequences obtained from Illumina HiSeqX at Macrogen Inc., Korea, were filtered by Trimmomatic 0.33 (Bolger et al. Citation2014) and de novo assembled and confirmed by Velvet 1.2.10 (Zerbino and Birney Citation2008), SOAPGapCloser 1.12 (Zhao et al. Citation2011), BWA 0.7.17 (Li et al. Citation2009), and SAMtools 1.9 (Li Citation2013). Geneious R11 11.1.5 (Biomatters Ltd, Auckland, New Zealand) was used for annotation based on that of N. davidi (NC_023823; Yu et al. Citation2014). ARWEN (Laslett and Canbäck Citation2008) was used to annotate tRNAs. DNA sample and specimen (95% ethanol) are deposited in InfoBoss Cyber Herbarium (IN; J. Park, INH-00013).
Mitochondrial genome length of N. heteropoda koreana is 15,558 bp (Genbank accession is MK907921) and its GC ratio is 33.0%. It contains 13 protein-coding genes, 2 ribosomal RNAs, and 22 transfer RNAs. Gene order of N. heteropoda koreana is identical to those of available 24 mitogenome in Atyidae including N. davidi. Length of tRNAs of N. heteropoda koreana ranges from 63 bp to 70 bp, smaller than those of N. davidi (Yu et al. Citation2014) and Caridina gracilipes (Xu et al. Citation2016; maximum lengths of tRNA are 78 bp and 83 bp, respectively). Control region is 674 bp long (A + T 82.0%), presumably corresponding to the single largest non-coding AT-rich region.
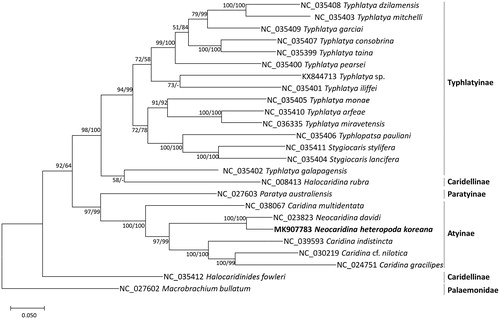
Thirteen protein-coding genes extracted from all available 24 mitochondrial genomes of family Atyidae including N. heteropoda koreana and one outgroup species (Macrobrachium bullatum; NC_027602) were aligned by MAFFT 7.388 (Katoh and Standley Citation2013) and concatenated. Bootstrapped Maximum likelihood and neighbor joining phylogenetic trees were constructed using MEGA X (Kumar et al. Citation2018). Phylogenetic trees show congruent relationship with (i) placement of Neocaridina within polyphyletic genus Caridina, (ii) polyphyly of genus Typhlatya, and (iii) Typhlatya galapagensis grouping with Halocaridina. However, incongruency of (i) placement of genus Halocaridinides and (ii) phylogenetic relationship among the subfamilies (von Rintelen et al. Citation2012; Jurado-Rivera et al. Citation2017) was also found (). Neocaridina heteropoda koreana mitochondrial genome will be utilized to investigate phylogenetic relationships of Neocaridina species.
Figure 1. Maximum likelihood (bootstrap repeat is 1,000), neighbor joining (bootstrap repeat is 10,000) phylogenetic trees of 24 Atyid mitochondrial genomes: Neocaridina heteropoda koreana (MK907921 in this study), Neocaridina davidi (NC_023823), Caridina multidentata (NC_038067), Caridina indistincta (NC_039593), Caridina cf. nilotica (NC_030219), Caridina gracilipes (NC_024751), Paratya australiensis (NC_027603), Halocaridina rubra (NC_008413), Typhlatya dzilamensis (NC_035402), Typhlatya mitchelli (NC_035403), Typhlatya garciai (NC_035409), Typhlatya consobrina (NC_035407), Typhlatya taina (NC_035399), Typhlatya pearsei (NC_035400), Typhlatya sp. (KX844713), Typhlatya iliffei (NC_035401), Typhlatya monae (NC_035405), Typhlatya arfeae (NC_035410), Typhlatya miravetensis (NC_036335), Typhlopatsa pauliani (NC_035406), Stygiocaris stylifera (NC_035411), Stygiocaris lancifera (NC_035404), Typhlatya galapagensis (NC_035402), Halocaridina rubra (NC_008413), Halocaridinides fowleri (NC_035412), and Macrobrachium bullatum (NC_027602) as an outgroup. Phylogenetic tree was drawn based on the maximum likelihood tree. The numbers above branches indicate bootstrap support values of maximum likelihood and neighbor-joining phylogenetic trees, respectively.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30:2114–2120.
- Crowl TA, McDowell WH, Covich AP, Johnson SL. 2001. Freshwater shrimp effects on detrital processing and nutrients in a tropical headwater stream. Ecology. 82:775–783.
- Jurado-Rivera JA, Pons J, Alvarez F, Botello A, Humphreys WF, Page TJ, Iliffe TM, Willassen E, Meland K, Juan C, Jaume D. 2017. Phylogenetic evidence that both ancient vicariance and dispersal have contributed to the biogeographic patterns of anchialine cave shrimps. Scientific Rep. 7:2852
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35:1547–1549.
- Laslett D, Canbäck B. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 24:172–175.
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv Preprint arXiv. 13033997.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25:2078–2079.
- Liang X. 2004. Fauna Sinica. Invertebrata: Crustacea: Decapoda: Atyidae. Beijing (China): Science Press (in Chinese).
- Mugo-Bundi J, Oyoo-Okoth E, Ngugi CC, Manguya-Lusega D, Rasowo J, Chepkirui‐Boit V, Opiyo M, Njiru J. 2015. Utilization of Caridina nilotica (Roux) meal as a protein ingredient in feeds for Nile tilapia (Oreochromis niloticus). Aquac Res. 46:346–357.
- Shih H-T, Cai Y, Niwa N, Nakahara Y. 2017. A new species of land-locked freshwater shrimp of the genus Neocaridina (Decapoda: Caridea: Atyidae) from Iki Island, Kyushu, Japan. Zool Stud. 56:1–14.
- Snyder MN, Small GE, Pringle CM. 2015. Diet‐switching by omnivorous freshwater shrimp diminishes differences in nutrient recycling rates and body stoichiometry across a food quality gradient. Freshwater Biol. 60:526–536.
- von Rintelen K, Page TJ, Cai Y, Roe K, Stelbrink B, Kuhajda BR, Iliffe TM, Hughes J, von Rintelen T. 2012. Drawn to the dark side: a molecular phylogeny of freshwater shrimps (Crustacea: Decapoda: Caridea: Atyidae) reveals frequent cave invasions and challenges current taxonomic hypotheses. Mol Phylogenet Evol. 63:82–96.
- Xu G, Du F, Nie Z, Xu P, Gu R. 2016. Complete mitochondrial genome of Caridina nilotica gracilipes. Mitochondrial DNA Part A. 27:1249–1250.
- Yu Y-Q, Yang W-J, Yang J-S. 2014. The complete mitogenome of the Chinese swamp shrimp Neocaridina denticulata sinensis Kemp 1918 (Crustacea: Decapoda: Atyidae). Mitochondrial DNA. 25:204–205.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829.
- Zhao Q-Y, Wang Y, Kong Y-M, Li X, Hao P. 2011. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinformatics. 12:S2.
