Abstract
We presented complete chloroplast genome of Coffea arabica BM1 which is 155,189 bp long and has four subregions: 85,159 bp of large single copy (LSC), and 18,135 bp of small single copy (SSC) regions are separated by 25,946 bp of inverted repeat (IR) regions including 131 genes (86 protein-coding genes, 8 rRNAs, and 37 tRNAs). The overall GC content of the chloroplast genome is 37.4% and those in the LSC, SSC, and IR regions are 35.4, 31.3, and 43.0%, respectively. Low level of cultivar specific sequence variations was found among seven coffee chloroplast genomes.
Coffea arabica is one of the major species for coffee production (Lashermes et al. Citation1999, Citation2009; O'brien and Kinnaird Citation2003). Cultivar Blue Mountain was introduced in Blue Mountains area of Jamaica in 1728 and now it has also been cultivated in Kenya, Hawaii, Haiti, Papua New Guinea, and Cameroon (Clifford Citation2012). Most of the Blue Mountain coffee was exported to Japan (Teuber Citation2010). It has unique mutations in comparison to Typica, presenting resistance to coffee berry disease (Clifford Citation2012). To investigate genetic diversity of coffee chloroplast genomes, we chose another cultivar, Blue Mountain which is similar to C. arabica ‘Typica’ (Min et al., accepted ) as a fifth coffee chloroplast genomes.
Total DNA of C. arabica ‘Blue Mountain’ (BM1; YS. Kim, IB-01024 in InfoBoss Cyber Herbarium (IN)) bought in Seoul (37.511566N, 127.059361E) was extracted from fresh leaves by using a DNeasy Plant Mini Kit (QIAGEN, Hilden, Germany). Genome sequencing was performed using HiSeq2000 at Macrogen Inc., Seoul, Korea, and raw sequences were filtered by Trimmomatic 0.33 (Bolger et al. Citation2014). de novo assembly and sequence confirmation were done by Velvet 1.2.10 (Zerbino and Birney Citation2008), SOAPGapCloser 1.12 (Zhao et al. Citation2011), BWA 0.7.17 (Li Citation2013), and SAMtools 1.9 (Li et al. Citation2009). Geneious R11 11.0.5 (Biomatters Ltd., Auckland, New Zealand) was used for chloroplast genome annotation based on Coffea arabica TY1 chloroplast genome (MK862266; Park et al., Citationunder review)
BM1 chloroplast genome (Genbank accession is MK875244) 155,189 bp long (GC ratio is 37.4%) and has four subregions: 85,159 bp of large single copy (35.4%) and 18,135 bp of small single copy (31.3%) regions are separated by 25,948 bp of inverted repeat (IR; 43.0%). It contains 131 genes (86 protein-coding genes, 8 rRNAs, and 37 tRNAs); 19 genes (8 protein-coding genes, 4 rRNAs, and 7 tRNAs) are duplicated in IR regions.
Based on alignment with six C. arabica chloroplast genomes (NC_008535 (Samson et al. Citation2007), KY085909, MK342634 (Park, Xi, et al. Citation2019), MK353209 (Park, Kim, Xi, Nho, et al. Citation2019), MK353212 (Park, Kim, Xi, Heo Citation2019), and MK862266 (Park et al. Citationunder review) named as CR, CA2, CH3, HP1, IN1, and TY1), three single nucleotide polymorphisms (SNPs) and six insertions and deletions (INDELs) against CR, one INDEL against CA2, six INDELs against CH3, 6 INDELs against HP1, 88 INDELs against IN1, and two INDELs against TY1 were found. It presents a low level of sequence variations on chloroplast genomes, similar to the cases of Marchantia polymorpha subsp. ruderalis (4 SNPs; Kwon et al. Citation2019) and Camellia japonica (one SNP and one INDEL; Park, Kim, Xi, Oh, et al. Citation2019).
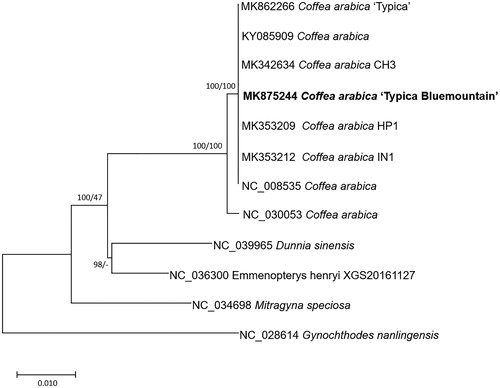
Seven Coffea and four chloroplast genomes of Rubiaceae as outgroup species were used for constructing neighbor-joining (bootstrap repeat is 10,000) and maximum likelihood (bootstrap repeat is 1000) trees using MEGA X (Kumar et al. Citation2018) based on alignment by MAFFT 7.388 (Katoh and Standley Citation2013). Phylogenetic trees show that BM1 is clustered with other Coffea arabica chloroplast genomes (), presenting a low level of variations on chloroplast genome among cultivars, reflecting homogeneity of C. arabica due to worldwide cultivation.
Figure 1. (A) Neighbor joining (bootstrap repeat is 10,000) and maximum likelihood (bootstrap repeat is 1000) phylogenetic trees of seven Coffea and four Rubiaceae complete chloroplast genomes: seven Coffea arabica (MK875244 in this study, MK862266, MK353212, NC_008535, KY085909, MK342634, and MK353209), Coffea canephora (NC_030053), Mitragyna speciosa (NC_034698), Dunnia sinensis (NC_039965), Emmenopterys henryi (NC_036300), and Gynochthodes nanlingensis (NC_028614). The numbers above branches indicate bootstrap support values of neighbor joining and maximum likelihood phylogenetic trees, respectively.
Disclosure statement
The authors declare that they have no competing interests.
Additional information
Funding
References
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30:2114–2120.
- Clifford MN. 2012. Coffee: botany, biochemistry and production of beans and beverage. Boston (MA): Springer.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35:1547–1549.
- Kwon W, Kim Y, Park J. 2019. The complete chloroplast genome of Korean Marchantia polymorpha subsp. ruderalis Bischl. & Boisselier: low genetic diversity between Korea and Japan. Mitochondr DNA B. 4:959–960.
- Lashermes P, Combes M-C, Robert J, Trouslot P, D'Hont A, Anthony F, Charrier A. 1999. Molecular characterisation and origin of the Coffea arabica L. genome. Mol General Genetics MGG. 261:259–266.
- Lashermes H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25:2078–2079.
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint arXiv. 1303.3997
- O'brien TG, Kinnaird MF. 2003. Caffeine and conservation. Science 300:587.
- Park J, Kim Y, Xi H, Heo K-I. 2019. The complete chloroplast genome of ornamental coffee tree, Coffea arabica L. (Rubiaceae). Mitochondr DNA B. 4:1059–1060.
- Park J, Kim Y, Xi H, Nho M, Woo J, Seo Y. 2019. The complete chloroplast genome of high production individual tree of Coffea arabica L. (Rubiaceae). Mitochondr DNA B. 4:1541–1542.
- Park J, Kim Y, Xi H, Oh YJ, Hahm KM, Ko J. 2019. The complete chloroplast genome of common camellia tree in Jeju island, Korea, Camellia japonica L. (Theaceae): intraspecies variations on common camellia chloroplast genomes. Mitochondr DNA B. 4:1292–1293.
- Park J, Xi H, Kim Y, Heo K-I, Nho M, Woo J, Seo Y, Yang JH. 2019. The complete chloroplast genome of cold hardiness individual of Coffea arabica L. (Rubiaceae). Mitochondr DNA B. 4:1083–1084.
- Park J, Kim Y, Xi H, Heo K-I. Under review. The complete chloroplast genome of coffee tree, Coffea arabica L. ‘Typica Bluemountain’ (Rubiaceae). doi:10.1080/23802359.2019.1636729
- Samson N, Bausher MG, Lee SB, Jansen RK, Daniell H. 2007. The complete nucleotide sequence of the coffee (Coffea arabica L.) chloroplast genome: organization and implications for biotechnology and phylogenetic relationships amongst angiosperms. Plant Biotechnol J. 5:339–353.
- Teuber R. 2010. Geographical indications of origin as a tool of product differentiation: The case of coffee. J Int Food Agribus Mark. 22:277–298.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829.
- Zhao Q-Y, Wang Y, Kong Y-M, Luo D, Li X, Hao P. 2011. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinformatics. 12:S2.
