Abstract
The fall armyworm, Spodoptera frugiperda, is a serious pest in large numbers on more than 350 plant species in the world. We have determined a 15,388 bp mitogenome of S. frugiperda which includes 13 protein-coding genes, two ribosomal RNA genes, and 22 transfer RNAs. The base composition was AT-biased (81.3%). Phylogenetic trees present that Korean S. frugiperda placed in basal position of S. frugiperda clade. S. frugiperda mitochondrial genome can be used for understanding recent active migration of S. frugiperda.
The fall armyworm, Spodoptera frugiperda, is one of the most important noctuid moth pests feeds in more than 350 plant species, damaging maize, rice, sorghum, sugarcane, wheat, vegetable crops, and cotton (Montezano et al. Citation2018). This species is native to tropical and subtropical regions of the Americas; however, it was first reported from Africa in 2016 (Goergen et al. Citation2016) and from the Indian subcontinent in 2018 (Ganiger et al. Citation2018). In Korea, the moth was first found at corn fields of Jocheon and Gujwa, Jeju, in early June, and subsequently at many counties of Jella-do and Gyeongsang-do in June and July in this year.
Genomic DNA of S. frugiperda collected on Zea mays L. in Hwayang-myeon, Yeosu-si, Jeollanam-do, Republic of Korea in 2019 (34°68′76ʺN, 127°58′17ʺE; the specimen is stored in Gyeongsang National University, Korea accession number: Coll#HB101) was extracted using DNeasy Brood &Tissue Kit (QIAGEN, Hilden, Germany). HiSeqX was used for sequencing (Macrogen Inc., Korea). Filtering, de novo assembly, and gap-filing processes were done by Velvet 1.2.10 (Zerbino and Birney Citation2008), Trimmomatic 0.33 (Bolger et al. Citation2014), SOAPGapCloser 1.12 (Zhao et al. Citation2011), BWA 0.7.17 (Li Citation2013), SAMtools 1.9 (Li et al. Citation2009). Geneious R11 11.1.5 (Biomatters Ltd, Auckland, New Zealand) and ARWEN (Laslett and Canbäck Citation2008) were used to annotate S. frugiperda based on alignment of other Spodoptera mitochondrial genomes.
S. frugiperda mitochondrial genome (Genbank accession is MN385596) is 15,388 bp long, the second longest among four S. frugiperda mitochondrial genomes. Its GC ratio is 18.7%, the lowest among four. It contains 13 protein-coding genes (PCGs), two rRNAs, and 22 tRNAs. Gene order of S. frugiperda is identical to other Spodoptera mitochondrial genomes, presenting conserved gene order of Noctuoidea mitochondrial genomes (Liu et al. Citation2016).
Based on alignments against three S. frugiperda mitochondrial genomes (NC_027836, KU877172, and KJ778892), 43 insertions or deletions (INDELs) and 275 single nucleotide polymorphisms (SNPs), 49 INDELs and 351 SNPs, and 74 INDELs and 247 SNPs were identified, respectively. Amounts of these intraspecies sequence variations are similar to that of Chilo suppressalis (79 SNPs and 291 INDELs) (Park, Xi, et al. Citation2019) and more than those of Laodelphax striatellus (140 and 40 SNPs and 166 and 118 INDELs) (Park, Jung, et al. Citation2019), Nilaparvata lugens (112 SNPs and 59 INDELs) (Park, Kwon, et al. Citation2019) and Aphis gossypii (61 SNPs and 3 INDELs; Park, Xi et al., Citation2019).
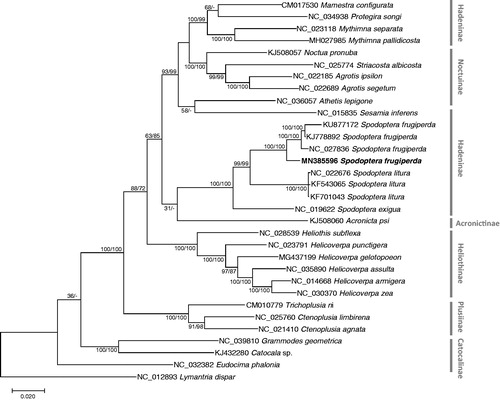
We inferred the phylogenetic relationship of 31 Noctuidae mitochondrial genomes including Spodoptera species and Lymantria dispar as an outgroup. Multiple sequence alignment was conducted by MAFFT 7.388 (Katoh and Standley Citation2013) using concatenated alignments of 13 PCGs. Bootstrapped maximum likelihood and neighbor joining trees were constructed using MEGA X (Kumar et al. Citation2018). Korean S. frugiperda is clustered with other three S. frugiperda (), and all mitogenomes of Spodoptera genus form one clade, presenting monophyletic manner (). Four mitogenomes of S. frugiperda showed intraspecies variations, suggesting the new mitogenome analyzed in this study will be helpful to understand geographical genetic variations of S. frugiperda.
Figure 1. Maximum likelihood (bootstrap repeat is 1000) and neighbor-joining (bootstrap repeat is 10,000) phylogenetic tree of all Spodoptera mitochondrial genomes: Spodoptera frugiperda (MN385596: this study, NC_027836, KJ778892, and KU877172), Spodoptera exigua (NC_019622), Spodoptera litura (NC_022676, KF701043, and KU877172), 23 Noctuidae species: Acronicta psi (KJ508060), Agrotis ipsilon (NC_022185), Agrotis segetum (NC_022689), Athetis lepigone (NC_036057), Catocala sp. (KJ432280), Ctenoplusia agnate (NC_021410), Ctenoplusia limbirena (NC_025760), Eudocima phalonia (NC_032382), Grammodes geometrica (NC_039810), Helicoverpa armigera (NC_014668), Helicoverpa assulta (NC_035890), Helicoverpa gelotopoeon MG437199), Helicoverpa punctigera (NC_023791), Helicoverpa zea (NC_030370), Heliothis subflexa (NC_028539), Mamestra configurata (CM017530), Mythimna pallidicosta (MH027985), Mythimna separata (NC_023118), Noctua pronuba (KJ508057), Protegira songi (NC_034938), Sesamia inferens (NC_015835), Striacosta albicosta (NC_025774), Trichoplusia ni (CM010779), and Lymantria dispar (NC_012893) as an outgroup. Phylogenetic tree was drawn based on maximum likelihood phylogenetic tree. The numbers above branches indicate bootstrap support values of maximum likelihood and neighbor joining phylogenetic trees, respectively.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120.
- Ganiger PC, Yeshwanth HM, Muralimohan K, Vinay N, Kumar ARV, Chandrashekara K. 2018. Occurrence of the new invasive pest, fall armyworm, Spodoptera frugiperda (J.E. Smith) (Lepidoptera: Noctuidae) in the maize fields of Karnataka, India. Current Science 115:621–623.
- Goergen G, Kumar PL, Sankung SB, Togola A, Tamò M. 2016. First report of outbreaks of the fall armyworm Spodoptera frugiperda (J E Smith) (Lepidoptera, Noctuidae), a New Alien Invasive Pest in West and Central Africa. PLoS One. 11(10):e0165632.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35:1547–1549.
- Laslett D, Canbäck B. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 24(2):172–175.
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint arXiv:13033997.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25(16):2078–2079.
- Liu Q-N, Chai X-Y, Bian D-D, Ge B-M, Zhou C-L, Tang B-P. 2016. The complete mitochondrial genome of fall armyworm Spodoptera frugiperda (Lepidoptera: Noctuidae). Genes Genom. 38(2):205–216.
- Montezano DG, Sosa-Gómez DR, Roque-Specht VF. 2018. Host plants of Spodoptera frugiperda (Lepidoptera: Noctuidae) in the Americas. Afr Entomol. 26:16.
- Park J, Jung JK, Ho Koh Y, Park J, Seo BY. 2019. The complete mitochondrial genome of Laodelphax striatellus (Fallén, 1826) (Hemiptera: Delphacidae) collected in a mid-Western part of Korean peninsula. Mitochondrial DNA Part B. 4(2):2229–2230.
- Park J, Kwon W, Park J, Kim H-J, Lee B-C, Kim Y, Choi NJ. 2019. The complete mitochondrial genome of Nilaparvata lugens (stål, 1854) captured in Korea (Hemiptera: Delphacidae). Mitochondrial DNA Part B. 4(1):1674–1676.
- Park J, Xi H, Kim Y, Park J & Lee W (2019) The complete mitochondrial genome of Aphis gossypii Glover, 1877 (Hemiptera: Aphididae) collected in Korean peninsula, Mitochondrial DNA Part B, 4(2):3007–3009.
- Park J, Xi H, Kwon W, Park C-G, Lee W. 2019. The complete mitochondrial genome sequence of Korean Chilo suppressalis (Walker, 1863) (Lepidoptera: Crambidae). Mitochondrial DNA Part B. 4(1):850–851.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18(5):821–829.
- Zhao QY, Wang Y, Kong YM, Luo D, Li X, Hao P. 2011. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinform. 12 Suppl 14:S2.
