Abstract
The complete plastid genome of Gentiana leucomelaena Maxim., belonging to the most species-rich section Chondrophyllae in Gentiana, was determined and analyzed in this study. It has a circular-mapping molecular with the length of 131,856 bp, the shortest one among all available Gentiana plastomes. Gentiana leucomelaena has gene mutation, for example ndh and rpl2 intron, and reversed SSC region comparing with the available species in sections Cruciata, Frigida, Kudoa, Isomeria and Microsperma. Phylogenetic analysis showed that G. leucomelaena clustered together with section Cruciata with a long branch. The plastome provides in this work will contribute to elucidate the phylogenetics and evolution in Gentiana.
Gentiana is an alpine group containing 362 species (Ho and Liu Citation2001), with the Qinghai-Tibet Plateau as the distribution and diversity center (Ho and Liu Citation2001; Favre et al., Citation2016). Among the 13 sections in Gentiana, section Chondrophyllae Bunge is the largest one containing about 180 species which are further divided into 12 series (Ho and Liu Citation2001; Favre et al. Citation2020). Gentiana leucomelaena Maxim., belonging to section Chondrophyllae series Humiles Marquand, has distribution range in Qinghai-Tibetan Plateau and adjacent areas. As the most species-rich section, there is very limited available plastome data in section Chondrophyllae at present (Fu et al. Citation2021).
Herein, we reported and characterized the complete plastome of G. leucomelaena (MT905404). One G. leucomelaena individual was collected from Jiangda, Tibet, China (31°38′N, 98°25′E). The voucher specimen was deposited at Herbarium of Luoyang Normal University (Bing Cai, [email protected]) under the voucher number Miao1902. The whole plant was used for DNA extraction using a Dzup plant genomic DNA extraction kit (Sangon, Shanghai, China). The fragmented genomic DNA was sequenced using Illumina HiSeq 2500 (Novogene, Tianjing, China), yielding 3 Gb of 150-bp paired-end reads. The plastome was de novo assembled by NOVOPlasty 2.6.1 (Dierckxsens et al. Citation2016) and annotated by GeSeq (Tillich et al. Citation2017) using the default parameters. Comparative analysis was conducted by mVISTA (Frazer et al. Citation2004) with species from five plastome-available Gentiana sections, Cruciata (Zhou et al. Citation2018), Frigida (She et al. Citation2019; Sun, Wang, et al. 2019), Kudoa and Isomeria (Fu et al. Citation2016; Sun et al. Citation2018) and Microsperma (Sun, Zhou, et al. 2019). Shared protein coding genes in available Gentiana plastomes were extracted and aligned by using MAFFT (Katoh et al. Citation2002). Using concatenated protein coding genes, maximum likelihood phylogenetic analyses were conducted with IQ-TREE (Nguyen et al. Citation2015) in PhyloSuite (Zhang et al. Citation2020) with 1000 replicates. Metagentiana rhodantha (MN822304) was served as the outgroup.
The complete G. leucomelaena plastome is a circular-mapping molecule with the length of 131,856 bp, the shortest in all available Gentiana plastomes. The LSC, IR and SSC regions were 75,476, 23,259 and 9862 bp, respectively. A total of 122 genes were annotated, containing 80 protein-coding genes, 34 tRNA genes and 8 rRNA genes. Comparison analysis indicated that plastome of G. leucomelaena has numerous gene losses such as ndh complex and rpl2 intron, which is similar with G. section Kudoa (Sun et al. Citation2018). In addition, the SSC region in G. leucomelaena plastome is reversed comparing with all other Gentiana plasomtes.
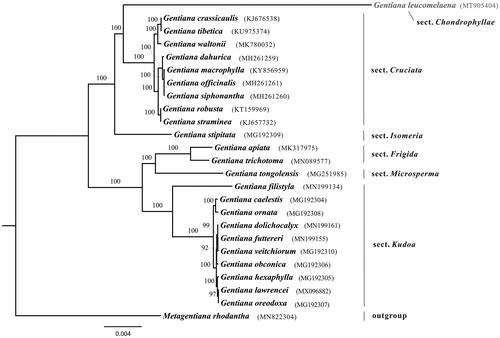
Comparison analysis showed that the hotspots among sections Chondrophyllae, Cruciata, Frigida, Isomeria, Kudoa and Microsperma located at intergenic regions, for instance trnH-GUG–psbA, atpH–atpI, petN–trnD, rbcL–accD and trnL-UAG–ccsA. The phylogenetic relationships among studied sections were fully supported. Phylogenetic analysis showed that G. leucomelaena was sister to section Cruciata with a long branch (), which was consistent with previous study (Favre et al. Citation2016). The determination of the G. leucomelaena plastome sequences provided new molecular data to illuminate the phylogenetics and molecular evolution of Gentiana.
Disclosure statement
All the authors declare no conflicts in this study.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MT905404. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA738973, SRR14859452, and SAMN19768589, respectively.
References
- Dierckxsens N, Mardulyn P, Smits G. 2016. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Favre A, Michalak I, Chen C‐H, Wang J‐C, Pringle JS, Matuszak S, Sun H, Yuan Y‐M, Struwe L, Muellner‐Riehl AN, et al. 2016. Out-of-Tibet: the spatio-temporal evolution of Gentiana (Gentianaceae). J Biogeogr. 43(10):1967–1978.
- Favre A, Pringle JS, Heckenhauer J, Kozuharova E, Gao Q, Lemmon EM, Lemmon AR, Sun H, Tkach N, Gebauer S, et al. 2020. Phylogenetic relationships and sectional delineation within Gentiana (Gentianaceae). Taxon. 69(6):1221–1238.
- Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. 2004. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 32(Web Server issue):W273–W279.
- Fu P-C, Sun S-S, Twyford AD, Li B-B, Zhou R-Q, Chen S-L, Gao Q-B, Favre A. 2021. Lineage-specific plastid degradation in subtribe Gentianinae (Gentianaceae). Ecol Evol. 11(7):3286–3299.
- Fu PC, Zhang YZ, Geng HM, Chen SL. 2016. Thse complete chloroplast genome sequence of Gentiana lawrencei var. farreri (Gentianaceae) and comparative analysis with its congeneric species. PeerJ. 4:e2540.
- Ho TN, Liu SW. 2001. A worldwide monograph of Gentiana. Beijing: Science Press.
- Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30(14):3059–3066.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- She RX, Li W, Xie X, Liu M, Zhao P. 2019. The complete nucleotide sequence of chloroplast genome of Gentiana apiata (Gentianaceae), an endemic medicinal herb in China. Mitochondrial DNA Part B. 4(2):2596–2597.
- Sun SS, Fu PC, Zhou XJ, Cheng YW, Zhang FQ, Chen SL, Gao QB. 2018. The complete plastome sequences of seven species in Gentiana sect. Kudoa (Gentianaceae): insights into plastid gene loss and molecular evolution. Front Plant Sci. 9:493.
- Sun SS, Wang H, Fu PC. 2019a. Complete plastid genome of Gentiana trichotoma (Gentianaceae) and phylogenetic analysis. Mitochondrial DNA Part B. 4(2):2775–2776.
- Sun SS, Zhou XJ, Li ZZ, Song HY, Long ZC, Fu PC. 2019b. Intra-individual heteroplasmy in the Gentiana tongolensis plastid genome (Gentianaceae). PeerJ. 7:e8025.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Zhang D, Gao F, Li WX, Jakovlić I, Zou H, Zhang J, Wang GT. 2020. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour. 20(1):348–355.
- Zhou T, Wang J, Jia Y, Li W, Xu F, Wang X. 2018. Comparative chloroplast genome analyses of species in Gentiana section Cruciata (Gentianaceae) and the development of authentication markers. Inter J Mol Sci. 19:1962.