Abstract
As a part of phylogenomic study of graminids, we report the complete plastome sequence of Flagellaria indica L. (Flagellariaceae) (NCBI No. MZ504969). This is the first reported complete plastome sequence from the Flagellariaceae. This plastome shows typical quadripartite structure. The plastome size is 161,643 bp, which consists of 88,714 bp large single copy (LSC), 19,065 bp small single-copy (SSC), and 26,932 bp inverted repeat (IR) regions. However, we detected F. indica plastome has a 288 bp small inversion between ycf3 and trnS-GGA. The palindromic repeats of 10 bp (TTCCAATTTC/GAAATTGGAA) were located on the two break points of inversion. F. indica plastome contains 113 genes, including 79 protein-coding, 30 tRNA, and four rRNA genes. Unlike other families of graminids, the functional ycf1 and ycf2 genes exist. Sixteen genes contain one intron and two genes (clpP and ycf3) have two introns. Sixty-two simple sequence repeat (SSR) loci are scattered in the plastome, respectively. The phylogenetic tree shows that Flagellariaceae are the basal sister lineage of other graminid families.
The graminid clade of Poales consist of four families; Flagellariaceae, Ecdeiocoleaceae, Joinvilleaceae, and Poaceae (APG IV Citation2016). As a part of phylogenomic study of graminids, we generated a complete plastome sequence of Flagellaria indica Linnaeus 1753 (Flagellariaceae) (NCBI No. MZ504969). This is the first reported complete plastome sequence from the family Flagellariaceae. The family consists of a single genus and five species (Christenhusz et al. Citation2018). Flagellaria indica is distributed in tropical and subtropical regions of Asia, East Africa, Australia, and Pacific islands. Other species mainly distribute in Pacific islands (Wepfer and Linder Citation2014).
The dried leaves of F. inidica were sampled from Koh Kong city, Koh Kong Province, Cambodia (N11°36′08.01″, E103°00′36.61″). They were ground into powder in liquid nitrogen and total DNAs were extracted using the G-spinTM IIp for Plant Genomic DNA Extraction Kit (iNtRON Biotechnology). The voucher specimen was deposited in the Korea University Herbarium (KUS acc. no. TCA2009-1388) and genomic DNA is deposited in the Plant DNA Bank in Korea (PDBK acc. no. 2009-1388). The complete plastome sequence was generated using an Illumina NovaSeq platform (Illumina Inc., San Diego, CA). De novo assembly was performed using the NOVOPlasty 4.3.1 (Dierckxsens et al. Citation2017). Annotation was performed using the Geneious 11.1.5 (Biomatters Ltd.; Kearse et al. Citation2012), National Center for Biotechnology Information (NCBI) BLAST, and tRNAscan-SE programs (Lowe and Eddy Citation1997). The average coverage of F. indica plastome was 464.9x. The simple sequence repeats (SSRs) were detected with the Phobos v. 3.3.12 program (Leese et al. Citation2008) in the Geneious 11.1.5. For the phylogenetic analysis, we selected and downloaded 11 related complete plastome sequences based on the APG IV system (APG IV Citation2016) from the NCBI database.
The plastome size of F. indica is 161,643 bp, which consists of 88,714 bp large single copy (LSC) region, 19,065 bp small single-copy region (SSC), and 26,932 bp inverted repeat (IR) region. This shows a typical quadripartite structure. However, we detected F. indica plastome has a 288 bp small inversion between ycf3 and trnS-GGA. The palindromic repeats of 10 bp (TTCCAATTTC/GAAATTGGAA) were located on the two break points of inversion. This inversion is shared by the graminids group in Poales (Hiratsuka et al. Citation1989; Ogihara et al. Citation2002; Wysocki et al. Citation2016). The plastome holds 113 unique genes, including 79 protein-coding genes, 30 tRNA genes, and 4 rRNA genes. Six protein-coding, eight tRNA, and four rRNA genes are duplicated in the IR regions. Unlike other families of graminids, the functional ycf1 and ycf2 genes exist (Darshetkar et al. Citation2019). The average A-T content of the plastome is 62.8%. Sixteen genes contain one intron and two genes, ycf3 and clpP, have two introns. A total of 62 simple sequence repeat (SSR) loci are distributed throughout the F. indica plastome. Among these, 40, 15, and 7 are mono-SSR, di-SSR, and tri-SSR loci, respectively.
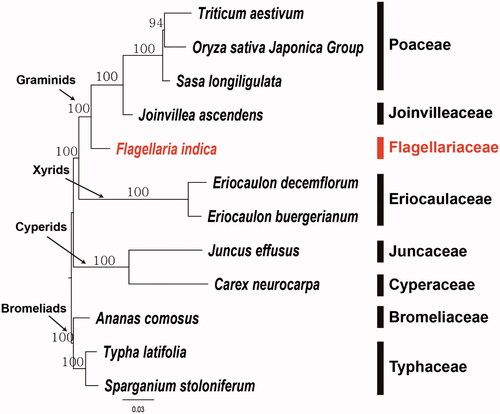
To establish the phylogenetic relationships of Flagellariaceae, we constructed a maximum likelihood tree using 12 Poales taxa. Phylogenetic analysis was performed on a data set that included 71 protein-coding genes and four rRNA genes from the 12 selected taxa using RAxML v.8.2.12 in CIPRES webserver (Stamatakis Citation2014) with a GTR + G + I model and 100 bootstrap replicates. The 75 gene sequences (59,754 bp in length) were aligned with the MUSCLE program using Geneious v. 11.1.5 (Biomatters Ltd.; Kearse Citation2012). The resulting tree supports the monophyly of graminids by 100% bootstrap value. Within the graminid clade, Flagellariaceae (F. indica) was a basal independent lineage and Joinvilleaceae and Poaceae from a subclade. The graminids was the sister group of xyrids (). Restiids was suggested as a sister group to the graminids by previous researchers (Bouchenak-Khelladi et al. Citation2014; Linder and Rudall Citation2005). However, none of complete plastome sequence of restiids are available at this moment. In contrast, abundant plastome sequences are available from Poaceae. In order to establishes the comprehensive phylogenetic relationships of Poales at family level, the plastome sequences from various unexploited small families are required. Therefore, the complete plastome sequence of F. indica (Flagellariaceae) will provide a useful resource for the evolutionary and phylogenomic studies of Poales.
Disclosure statement
The authors report no conflicts of interest for the content and writing of the paper.
Data availability statement
A specimen was deposited at the Korea University Herbarium (KUS, http://mpl.korea.ac.kr/herbarium.asp, Ki-Joong Kim, [email protected]) under the voucher number TCA2009-1388 and genomic DNA is deposited in the Plant DNA Bank in Korea (PDBK, http://pdbk.korea.ac.kr/index.asp, Ki-Joong Kim, [email protected] under the DNA no. PDBK 2009-1388. The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov under the accession no. MZ504969 for Flagellaria indica. The associated BioProject, BioSample, and SRA numbers are PRJNA749810, SAMN20420846, and SRR15253612, respectively.
Additional information
Funding
References
- APG IV. 2016. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot J Linn Soc. 181(1):1–20.
- Bouchenak-Khelladi Y, Muasya AM, Linder HP. 2014. A revised evolutionary history of Poales: origins and diversification. Bot J Linn Soc. 175(1):4–16.
- Christenhusz MJM, Fay MF, Chase MW. 2018. Plants of the World: an illustrated encyclopedia of vascular plants. Richmond: Kew Publishing.
- Darshetkar AM, Datar MN, Tamhankar S, Li P, Choudhary RK. 2019. Understanding evolution in Poales: insights from Eriocaulaceae plastome. PLOS One. 14(8):e0221423.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Hiratsuka J, Shimada H, Whittier R, Ishibashi T, Sakamoto M, Mori M, Kondo C, Honji Y, Sun CR, Meng BY. 1989. The complete sequence of the rice (Oryza sativa) chloroplast genome: intermolecular recombination between distinct tRNA genes accounts for a major plastid DNA inversion during the evolution of the cereals. Mol Gen Genet. 217(2–3):185–194.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Leese F, Mayer C, Held C. 2008. Isolation of microsatellites from unknown genomes using known genomes as enrichment templates. Limnol Oceanogr Methods. 6(9):412–426.
- Linder HP, Rudall PJ. 2005. Evolutionary history of Poales. Annu Rev Ecol Evol Syst. 36(1):107–124.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25(5):955–964.
- Ogihara Y, Isono K, Kojima T, Endo A, Hanaoka M, Shiina T, Terachi T, Utsugi S, Murata M, Mori N, et al. 2002. Structural features of a wheat plastome as revealed by complete sequencing of chloroplast DNA. Mol Genet Genomics. 266(5):740–746.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Wepfer PH, Linder HP. 2014. The taxonomy of Flagellaria (Flagellariaceae). Aust Syst Bot. 27(3):159–179.
- Wysocki WP, Burke SV, Swingley WD, Duvall MR. 2016. The first complete plastid genome from Joinvilleaceae (J. ascendens; Poales) shows unique and unpredicted rearrangements. PLOS One. 11(9):e0163218.