Abstract
Flemingia macrophylla (Willd.) Prain is an ethnomedicinal plant with high nutritional and medicinal values. In this study, we report the complete chloroplast genome of F. macrophylla. The chloroplast genome has a typical quadripartite structure with a genome size of 152,988 bp, including a large single-copy (LSC) of 83,634 bp, a small single-copy (SSC) of 17,774 bp and two inverted repeats (IRs) of 25,790 bp. The genome contains 129 genes, including 84 protein-coding, 37 tRNA and 8 rRNA genes. The overall GC content is 35.1%. Phylogenetic analysis showed that F. macrophylla grouped with a clade containing the genera of Fagelia, Dolichos, Eriosema, Dunbaria and Cajanus in Fabaceae. This study provides essential data and insight for understanding the phylogenetic placement of Flemingia.
The root of Flemingia macrophylla (Willd.) Prain 1897 classified in the Fabaceae (Xu et al. Citation2010) are used in traditional medicine as documented in the Pharmacopeia of the People’s Republic of China (Volume I, 2015 Edition) (Chinese Pharmacopoeia Commission Citation2015). The plant has also been widely used as ethnomedicine and for a diet to treat rheumatic bone pain, lumbar muscle strain, Kala-azar, fever, etc (Rana et al. Citation2011). So far, there has been no genome-scale (phylogenetic) study of Flemingia. As plastid genomes have been widely applied for phylogenetic reconstruction, species identification, population genetics and selection test (Mehmood, Abdullah Ubaid Z, Bao, et al. Citation2020, Mehmood, Abdullah Ubaid Z, Shahzadi, et al. Citation2020). In this study, we report the complete chloroplast genome sequence of F. macrophylla and its phylogenetic relationship to closely related genera in Papilionoideae.
Total genomic DNA was extracted from the silica-dried leaves of F. macrophylla using the CTAB method (Doyle Citation1987), which were collected from a transplanted individual in Guilin Botanical Garden (25.0704 N, 110.2991 E). The voucher specimen was deposited at the Herbarium of Guangxi Institute of Botany (http://www.gxib.cn/spIBK/, Contact person name: Chun-Rui Lin, Email: [email protected]) under the voucher number IBK00432997. The high throughput genomic sequencing with paired ends (PE150) was performed on a NovaSeq 6000 (in Novogene corp., Tianjin, China). Approximately 3 Gb of clean data was obtained after quality filtering using fastp (Chen et al. Citation2018). The chloroplast genome of F. macrophylla was assembled with default settings using SPAdes 3.11.0 (Bankevich et al. Citation2012) and annotated using PGA (Qu et al. Citation2019). Analysis of the boundaries between IRs and single-copy regions was performed by using the online program IRSCOPE (Amiryousefi et al. Citation2018). The average coverage depth was calculated by mapping all the raw reads without trimming to the de novo assembled chloroplast genome in BWA-MEM (Li Citation2013) and SAMtools (Danecek et al. Citation2021). The complete chloroplast genome sequence of F. macrophylla was submitted to GenBank (accession number: MZ274347).
The calculated average coverage depth of F. macrophylla is 507 X. The chloroplast genome has a typical quadripartite structure, with a total length of 152,988 bp and an overall GC content of 35.1%, which contains one LSC region (83,634 bp), one SSC region (17,774 bp) and two IR regions (25,790 bp, respectively). It contains 129 genes, including 84 protein-coding, 8 rRNA and 37 tRNA genes. Among them, 15 genes contain one intron, and two genes contain two introns. Analysis of the boundaries between the IRs and single-copy regions of F. macrophylla indicated that the rps19 gene spans the LSC/IRb boundary with a length of 230 bp in the LSC and 49 bp in the IRb; the ycf1 gene spans the SSC/IRa boundary with a length of 4848 bp in the SSC and 495 bp in the IRa; a pseudogene (ψycf1) lies at the IRb/SSC boundary, and the trnH gene is 30 bp away from the LSC/IRa junction. The structure of this chloroplast genome is generally in line with others of Papilionoideae reported, with a few minor differences, such as for the location of the rps19 gene, which spans the LSC and IRb region in F. macrophylla but is wholly contained in the LSC region in most other species of Papilionoideae that have been reported (Zha et al. Citation2020).
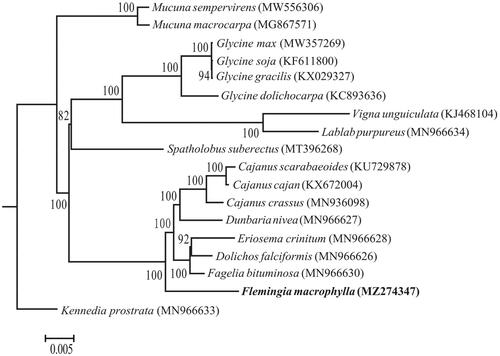
To reveal the phylogenetic position of F. macrophylla, a maximum likelihood tree was reconstructed using RAxML (Stamatakis Citation2014) with the GTR + GAMMA substitution model based on the concatenated data of 77 protein-coding genes from the chloroplast genome sequences of 18 species. Kennedia prostrata was designated as the outgroup, and the tree was evaluated based on 1,000 bootstrap replicates. The phylogenetic analysis fully resolved the phylogeny and indicated that F. macrophylla grouped with a clade containing Fagelia bituminosa, Dolichos falciformis, Eriosema crinitum, Dunbaria nivea, Cajanus crassus, Cajanus cajan and Cajanus scarabaeoides (). This is consistent with other studies based on nuclear genes from transcriptomes and/or genomes of 333 genera of Fabaceae, in which F. macrophylla was also suggested to be closely related to a clade consisting of Dolichos, Dunbaria, Cajanus and Rhynchosia (Zhao et al. Citation2021).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that supports the findings of this study are openly available in GenBank at (https://www.ncbi.nlm.nih.gov/) under the accession no. MZ274347. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA732804, SRR14663578, and SAMN19341677, respectively.
Additional information
Funding
References
- Amiryousefi A, Hyvönen J, Poczai P. 2018. IRscope: an online program to visualize the junction sites of chloroplast genomes. Bioinformatics. 34(17):3030–3031.
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Chen S, Zhou Y, Chen Y, Jia G. 2018. Fastp: an ultra-fast all-in-one fastq preprocessor. Bioinformatics. 34(17):i884–i890.
- Chinese Pharmacopoeia Commission. 2015. Pharmacopoeia of the People’s Republic of China. Vol. I. Beijing: China Medical Science Press; p. 418.
- Danecek P, Bonfield JK, Liddle J, Marshall J, Ohan V, Pollard MO, Whitwham A, Keane T, McCarthy SA, Davies RM, et al. 2021. Twelve years of SAMtools and BCFtools. GigaScience. 10(2):1–4.
- Doyle JJ. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv: 1303.3997v2 [q-bio.GN].
- Mehmood F, Abdullah Ubaid Z, Bao Y, Poczai P, Mirza B. 2020. Comparative plastomics of Ashwagandha (Withania, Solanaceae) and identification of mutational hotspots for barcoding medicinal plants. Plants. 9(752):1–20.
- Mehmood F, Abdullah Ubaid Z, Shahzadi I, Ahmed I, Waheed MT, Poczai P, Mirza B. 2020. Plastid genomics of Nicotiana (Solanaceae): insights into molecular evolution, positive selection and the origin of the maternal genome of Aztec tobacco (Nicotiana rustica). PeerJ. 8:e9552.
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15:50.
- Rana Y, Rana VK, Jain VK, Jain B. 2011. Acute toxicity study and analgesic activity of successive extracts of roots of Flemingia macrophylla (Willd.). Int J Pharmacol Res. 2(2):62–67.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Xu LR, Chen DZ, Zhu XY, Huang PH, Wei Z, Sa R, Zhang DX, Bao BJ, Wu DL, Sun H, Gao XF, Liu YX, Chang ZY, Li JQ, Zhang ML, Podlech D, Ohashi H, Larsen K, Welsh SL, Vincent MA, Gilbert MG, Pedley L, Schrire BD, Yakovlev GP, Thulin M, Nielsen IC, Choi B, Turland NJ, Polhill RM, Larsen SS, Hou D, Iokawa Y, Wilmot-Dear CM, Kenicer G, Nemoto T, Lock JM, Salinas AD, Kramina TE, Brach AR, Bartholomew B, Sokoloff DD. 2010. Flora of China. In: Fabaceae (Leguminosae). Vol. 10. Beijing: Science Press; p. 235–236.
- Zha X, Wang XY, Li JR, Gao F, Zhou YJ. 2020. Complete chloroplast genome of sophora alopecuroides (papilionoideae): molecular structures, comparative genome analysis and phylogenetic analysis. J Genet. 99(1):13.
- Zhao Y, Zhang R, Jiang KW, Qi J, Hu Y, Guo J, Zhu R, Zhang T, Egan AN, Yi TS, et al. 2021. Nuclear phylotranscriptomics and phylogenomics support numerous polyploidization events and hypotheses for the evolution of rhizobial nitrogen-fixing symbiosis in Fabaceae. Mol Plant. 14(5):748–773.