Abstract
Secale strictum subsp. kuprijanovii is a perennial, hermaphrodite wild rye species and a progenitor of the modern cultivated rye, Secale cereale. With high adaptive capacity in stress conditions, it is valuable for enriching the germplasm resources of rye. Therefore, to elucidate its genetic and phylogenetic relationship is of great importance. We hereby sequenced, assembled and presented for the first time the complete chloroplast genome of this less studied species. The whole genome is 137,079 bp in size, including a large single copy region of 81,099 bp, a small single copy region of 12,820 bp and two separated inverted repeat regions of 43,160 bp. A total of 109 unique genes were annotated, including 67 protein-coding genes, 38 tRNA genes and 4 rRNA genes. Phylogenetic analysis showed that Secale strictum subsp. kuprijanovii clustered most closely with Secale cereal. A remarkably close evolutionary relationship of S.strictum subsp. kuprijanovii with various wheat varieties may indicate its usage as a genetic resource for the breeding of both the cultivated rye and wheat.
Secale is a genus under the subtribe Hordeinae of grass tribe Triticeae, in which crops like barley and wheat are also included. The probably most well-known species in Secale genus is the cultivated rye Secale cereal, grown as an annual grain and forage. Cereal rye and its wild forms provide important genetic resources for wheat breeding due to their excellent resistance to different stress (Singh et al. Citation2021). Secale strictum subsp. kuprijanovii, a perennial and hermaphrodite wild rye species with numerous small edible seeds born from caryopsis fruits, is recognized as an ancestral form of cultivated rye today proposed in common hypotheses (Frederiksen and Petersen Citation1998). Originally occurred in the Near East region and Armenia, its high adaptive potential and novel gene resources should be considered as reservoirs for breeding programs like many other Middle Eastern landrace populations (Hagenblad et al. Citation2016).
Previous SSR analysis and a nuclear region sequence diversity analysis of phylogenetic relationship in Secale genus demonstrated a clear separation between perennial S. strictum subspecies and the annual taxa (Maraci et al. Citation2018). A certain number of partial sequences of functional genes or regions in organelles of S. strictum subsp. kuprijanovii were publicated in NCBI nucleotide database, which contributed to indicating its phylogenetic position in the Secale genus (Ren et al. Citation2011; Evtushenko et al. Citation2017). However, neither the complete nuclear genome nor organelle genome sequence of S. strictum subsp. kuprijanovii has been published yet. The chloroplast genome, known to be conservative generally in land plants, can be crucial for phylogeny analysis (Sugiura Citation2003). Therefore, we sequenced and assembled the complete chloroplast genome of this wild kinship to rye, S. strictum subsp. kuprijanovii, in this study. The seeds of plants were harvested in Armenia (4°32′2″ N, 75°40.867′ W), dried and a specimen was deposited in the Herbarium of Zhejiang University under the voucher number HZU60244004 (contact person: Qinjie Chu; email: [email protected]). Fresh seedlings were then collected for total genomic DNA extraction using a modification of the CTAB method, and later sequenced by MGI platform (DNBSEQ-500) with a pair-end read length of 150 bp.
After quality control with NGSQCToolkit v2.3 (Patel and Jain Citation2012), the clean data was assembled by NOVOPlasty v3.6 (Dierckxsens et al., Citation2017), using complete chloroplast genome of Triticum aestivum (GenBank accession number NC_002762) as a reference. Genome annotation was performed by the GeSeq online (Tillich et al. Citation2017). The assembled genome sequences and annotation information have been submitted to DDBJ with an accession number LC645210. The chloroplast genome of S. strictum subsp. kuprijanovii has a total size of 137,079 bp, in which a distinct quadripartite structure exhibited, including a small single-copy region (SSC, 12,820bp), a large single-copy region (LSC, 81,099bp) and a pair of inverted repeats (IRa and IRb, 21,580 bp each). The GC contents in the whole genome, SSC, LSC, and IR regions are 38.2%, 32.1%, 36.2% and 43.9%, respectively. 109 unique genes were annotated altogether, including 67 protein-coding genes, 38 tRNA genes and 4 rRNA genes. A total of 21 genes contained a single intron, and only one gene (ycf3) contained two introns.
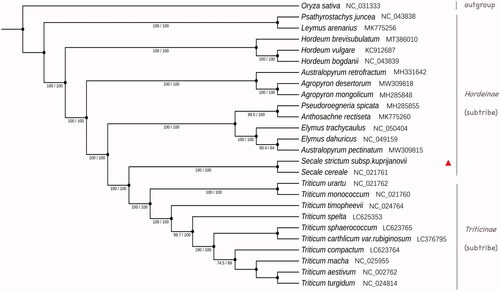
To illustrate more clearly its evolutionary position within the tribe Triticeae, we used complete chloroplast genome sequences of 26 species to construct a phylogenetic tree, including 13 perennial species from 9 genera in Hordeinae, 10 wheat varieties, 2 Secale species of S. strictum subsp. kuprijanovii and S. cereale plus Oryza sativa as an outgroup. Multiple sequence alignment was performed by MAFFT v7.475 (Katoh et al. Citation2002) with default parameters. The alignment results were calculated using the maximum likelihood algorithm and the phylogenetic tree was constructed by IQ-tree v1.6.12 (Nguyen et al. Citation2015) with parameters configured as ‘-st DNA -bb 1000 -alrt 1000.’ iTOL (Letunic and Bork Citation2019) was used to modify the constructed tree. The phylogenetic results first showed that S. strictum subsp. kuprijanovii clustered with S.cereale, and they diverged from the genus Triticum at one peculiar node (). The close evolutionary relationship of S. strictum subsp. kuprijanovii with the Triticum genus again implies this species being an abundant gene pool useful for improvement and breeding of both the cultivated rye and wheat.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI (https://www.ncbi.nlm.nih.gov/) under the accession number LC645210, and the associated BioProject, SRA, and Bio-Sample numbers are PRJNA749905, SRR15373215 and SAMN20425418 respectively.
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: De novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Evtushenko EV, Elisafenko EA, Gatzkaya SS, Lipikhina YA, Houben A, Vershinin AV. 2017. Conserved molecular structure of the centromeric histone CENH3 in Secale and its phylogenetic relationships. Sci Rep. 7(1):17628.
- Frederiksen S, Petersen G. 1998. A taxonomic revision of Secale (Triticeae, Poaceae). Nord J Bot. 18(4):399–420.
- Hagenblad J, Oliveira HR, Forsberg NEG, Leino MW. 2016. Geographical distribution of genetic diversity in Secale landrace and wild accessions. BMC Plant Biol. 16(1):1–20.
- Katoh K, Misawa K, Kuma KI, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30(14):3059–3066.
- Letunic I, Bork P. 2019. Interactive Tree of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47(W1):W256–W259.
- Maraci Ö, Özkan H, Bilgin R. 2018. Phylogeny and genetic structure in the genus Secale. PLoS One. 13(7):e0200825.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Patel RK, Jain M. 2012. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS One. 7(2):e30619.
- Ren TH, Chen F, Zou YT, Jia YH, Zhang HQ, Yan BJ, Ren ZL. 2011. Evolutionary trends of microsatellites during the speciation process and phylogenetic relationships within the genus Secale. Genome. 54(4):316–326.
- Singh AK, Zhang P, Dong C, Li J, Singh S, Trethowan R, Sharp P. 2021. Generation and molecular marker and cytological characterization of wheat - Secale strictum subsp. anatolicum derivatives . Genome. 64(1):29–38.
- Sugiura M. 2003. History of chloroplast genomics. Photosynth Res. 76(1–3):371–377.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.