Abstract
Acer leipoense is a rare and endangered species of the Sapindaceae with a very restricted distribution in Sichuan, China. In this study, the complete chloroplast genome of A. leipoense was characterized by de novo assembly using high-throughput sequencing. The chloroplast genome was 155,702 bp in length; it contained a large single copy region (85,890 bp) and a small single copy region (18,100 bp), which were separated by a pair of 25,856-bp inverted repeat regions. A total of 128 genes were predicted, including 83 protein-coding genes, 37 tRNA genes, and eight rRNA genes. A phylogenetic analysis of 23 chloroplast genome sequences from the genus Acer revealed that A. leipoense was closely related to A. yangbiense.
Acer L. (family Sapindaceae) is one of the most diverse tree genera in the Northern Hemisphere; it contains more than 150 species, most of which are found in eastern Asia (Li et al. Citation2019). Acer leipoense Fang and Soong is an endemic species in southwestern China that has significant ornamental value. The natural distribution of A. leipoense is restricted to Leibo county, Ebian county and Tianquan county in Sichuan Province, China, where it is scattered in subtropical mixed forests at altitudes of 1900–2700 m (Fang Citation1966; Xu et al. Citation2008, and our investigations in 2018–2020). Due to over-harvesting and habitat degradation, A. leipoense is now rare and has been included in the Threatened Species List of China’s Higher Plants in the endangered (EN) category (Qin et al. Citation2017). To date, the chloroplast genome of A. leipoense has not been sequenced, and little is known about the phylogenetic relationships of this species. In this study, we assembled the complete chloroplast genome of A. leipoense using next-generation sequencing, providing a genetic resource for further genetic and conservation studies.
Fresh leaves of A. leipoense were collected from Erlang Mountian, Tianquan County, Sichuan province, China (29°59'19.6″ N, 102°38'34.1″E). The field study complied with national Wild Plant Protective Regulations and we were allowed by the Tianquan Forestry Bureau to collect the required samples of plant material. A specimen (voucher number: BJFUZLC055) was deposited at the Museum of Beijing Forestry University (Liangcheng Zhao, [email protected]). Total genomic DNA was extracted with a modified CTAB method (Doyle and Doyle Citation1987), and paired-end sequencing was performed on the Illumina HiSeq 2000 platform to obtain approximately 2.5 Gbp of high-quality A. leipoense reads. Genome assembly was performed using GetOrganelle v1.6.2.0 (Jin et al. Citation2020) with k-mer length 95, and all predicted genes were annotated using PGA (Plastid Genome Annotator) software (Qu et al. Citation2019) with the Acer cesium Wall. ex Brandis chloroplast genome (MN315269) as a reference. The complete annotated chloroplast genome of A. leipoense was deposited at GenBank under the accession number MZ725939.
The Acer leipoense chloroplast genome displays a typical quadripartite organization: one large single copy region (LSC) of 85,890 bp and one small single copy region (SSC) of 18,100 bp separated by two inverted repeats (IRs) of 25,856 bp each. The total length of the chloroplast genome is 155,702 bp; it encodes 128 genes, including 83 protein-coding genes, 37 transfer RNA (tRNA) genes, and eight ribosomal RNA (rRNA) genes. Among these genes, 15 (atpF, ndhA, ndhB, petB, petD, rpl16, rpl2, rpoC1, rps16, trnA-UGC, trnG-UCC, trnI-GAU, trnK-UUU, trnL-UAA, and trnV-UAC) contain one intron and three (clpP, rps12, and ycf3) have two introns. The total GC content of the chloroplast genome is 37.9%, and the GC contents of the LSC, SSC, and IR regions are 36.1%, 32.2%, and 43.0%, respectively.
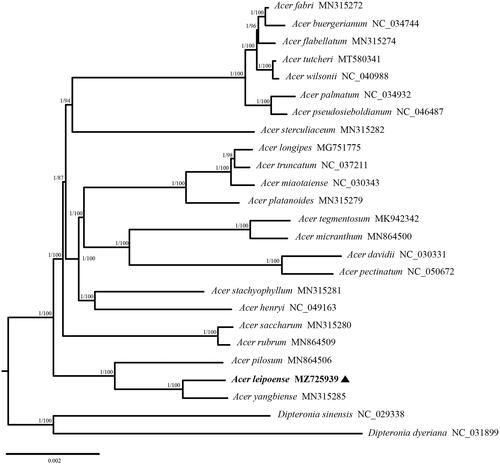
To determine the phylogenetic position of A. leipoense, 23 complete chloroplast genome sequences (one newly generated and 22 obtained from GenBank) from Acer and two outgroup sequences from Dipteronia were used for phylogenomic analyses. The sequences were aligned with MAFFT (Katoh et al. Citation2019), and ambiguously aligned regions were trimmed with Gblocks v0.91b (Castresana Citation2000). Maximum likelihood (ML) trees were inferred with the best model (GTR + F + R4, identified with ModelFinder) using IQ-TREE (Nguyen et al. Citation2015), and branch support values were obtained from 1000 bootstrap replicates. Bayesian inference (BI) was performed with MrBayes v3.2.3 (Ronquist et al. Citation2012) using the GTR + G + I model; it was set to run for four million generations with four chains, sampled every 1000 generations, with 25% of the trees discarded as burn-in. Analysis was run to completion and the average standard deviation of the split frequencies was < 0.01. BI and ML analyses recovered the same tree topology (). The results showed that A. leipoense was sister to A. yangbiense Chen and Yang with high support, consistent with morphological findings for the two species (Chen et al. Citation2003). A. leipoense, A. yangbiense, and A. pilosum Maxim. (1880) formed a strongly supported monophyletic group, which was located at the base of the clade formed by the other 20 Acer species.
Author contributions
LCZ and QYL conceived and designed the research; QYL, AW and YWG collected the samples, performed the experiments and analyzed the data; QYL and LCZ wrote the drafting of the paper and revised the manuscript. All authors approved the final manuscript and agreed to be accountable for all aspects of the work.
Disclosure statement
The authors declare there is no conflicts of interest and are responsible for the content.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. MZ725939. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA752922, SRR15372360, and SAMN20667766, respectively.
Additional information
Funding
References
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17: 540–552.
- Chen YS, Yang QE, Zhu GH. 2003. Acer yangbiense (Aceraceae), a new species from Yunnan, China. Novon. 13(3):296–299.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small amounts of fresh leaf tissue. Phytochem Bull. 19(1):11–15.
- Fang WP. 1966. Revisio taxorum Aceracearum sinicarum. Acta Phytotax. Sin. 11(2):139–189.
- Jin JJ, Yu WB, Yang JB, Song Y, de Pamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241.
- Katoh K, Rozewicki J, Yamada KD. 2019. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 20(4):1160–1166.
- Li J, Stukel M, Bussies P, Skinner K, Lemmon AR, Lemmon EM, Brown K, Bekmetjev A, Swenson NG. 2019. Maple phylogeny and biogeography inferred from phylogenomic data. Jnl of Sytematics Evolution. 57(6):594–606.
- Nguyen L, Schmidt HA, Von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Qin H, Yang Y, Dong S, He Q, Jia Y, Zhao L, Yu S, Liu H, Liu B, Yan Y, et al. 2017. Threatened species list of China’s higher plants. Biodivers Sci. 25(7):696–744.
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15:50.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP, et al. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Xu TZ, Chen YS, de Jong PC, Oterdoom HJ, Chang CS. 2008. Aceraceae. In: Wu ZY, Raven PH, editors. Flora of China. Vol. 11. Beijing: Science Press and Missouri Botanical Garden Press.