Abstract
Lepista sordida is a fairy ring-forming fungus that belongs to the family Tricholomataceae and is widely distributed in the Northern Hemisphere. Here, we report the complete mitochondrial genome sequence of L. sordida. The mitochondrial genome (57,375 bp) contained 20 protein-coding genes, 2 ribosomal RNA genes, and 26 transfer RNA genes. Phylogenetic analysis based on 14 conserved protein sequences from L. sordida and 15 related basidiomycetes showed that L. sordida was located on the outermost branch of the Tricholomataceae clade. This study is the first to report the complete mitochondrial genome sequence of a fairy ring-forming fungus belonging to the genus Lepista.
Lepista sordida (Schumach.) Singer 1951 is a mushroom belonging to the family Tricholomataceae (Kirk et al. Citation2008) and forms a circular pattern known as ‘fairy ring’ mainly on turfgrass, which is a phenomenon caused by the interaction between basidiomycete fungi and plants. To date, three compounds have been discovered and named as ‘fairy chemicals (FCs),’ which are involved in the formation of fairy rings (Choi et al. Citation2010a, Citation2010b, Citation2014; Kawagishi Citation2018). Genome analysis of L. sordida has revealed the biosynthetic pathway of FCs in this mushroom (Suzuki et al. Citation2016), and its genomic information is available on the web database F-RINGS (http://bioinf.mind.meiji.ac.jp/f-rings/) (Takano et al. Citation2019).
Mushrooms belonging to the genus Lepista are distributed worldwide and comprise approximately 50 species (Kirk et al. Citation2008); however, their limited morphological characteristics make it difficult to identify the species (Wang et al. Citation2019). In a previous study, the genus Lepista was divided into three clades by phylogenetic analysis based on multiple genes encoded in the nuclear genome (Alvarado et al. Citation2015). Comparative genome analysis and phylogenetic analysis of fairy ring-forming fungi, including L. sordida, will lead to a better understanding of the biological evolution of these fungi. However, it is not possible to confirm the results of phylogenetic analysis using the nuclear genome as there is no report of the complete mitochondrial sequence of the genus Lepista. In the present study, we determined the complete mitochondrial genome sequence of L. sordida and analyzed its phylogenetic relationship with related mushrooms.
Lepista sordida, which was originally collected from a lawn in Akita, Japan (40.2368 N, 140.5902 E), was deposited at Biological Resource Center (NBRC) of the National Institute of Technology and Evaluation (https://www.nite.go.jp/en/index.html, Culture Collection Division, [email protected]) under voucher no. NBRC 112841. The mycelia of L. sordida were cultivated in potato dextrose broth with 0.5% yeast extract at 25 °C for 2 weeks, and genomic DNA was extracted using the cetyltrimethylammonium bromide (CTAB) method as previously described (Tanaka et al. Citation2017). A genomic DNA library was constructed using the TruSeq DNA PCR-Free Sample Preparation Kit, and 301 bp paired-end sequencing was performed using the MiSeq System (Illumina, San Diego, CA). Raw sequence reads were cleaned using Trimmomatic ver. 0.36 (Bolger et al. Citation2014) by trimming adapter sequences and low-quality ends (quality score, <15), and reads with a high k-mer coverage (>200) were extracted using Khmer ver. 2.0 (Crusoe et al., Citation2015). The resulting 677,450 read pairs totaling approximately 373.2 Mb were assembled using NOVOPlasty ver. 4.0 (Dierckxsens et al. Citation2017) with the nucleotide sequence of the cytochrome c oxidase subunit 1 gene from Tricholoma flavovirens (GenBank accession no. NC_046501.1 nt. 1–1584) as the seed sequence. The mitochondrial genome of L. sordida consisted of a circular DNA molecule of 57,375 bp in length with a GC content of 26.0%.
Gene prediction and annotation of the mitochondrial genome of L. sordida were performed using MFannot ver. 1.36 (http://megasun.bch.umontreal.ca/cgi-bin/mfannot/mfannotInterface.pl) and curated manually. A large subunit ribosomal RNA gene was predicted by alignment to the mitochondrial genome of Coprinopsis cinerea (NW_003307477.1, CC1G_23001) using Geneious Prime 2021 (Kearse et al. Citation2012). The mitochondrial genome of L. sordida contained 48 genes, including 20 protein-coding genes, 2 rRNA genes (rnl and rns), and 26 tRNA genes. Overall, 20 protein-coding genes encoded 14 conserved mitochondrial proteins (cox1–3, cob, nad1–6, nad4L, atp6, atp8, and atp9), ribosomal protein S3, and 5 hypothetical proteins, 2 of which showed similarities to RNA polymerase and LAGLIDADG homing endonuclease. Only a large subunit ribosomal RNA gene harbored an intron of 488 bp.
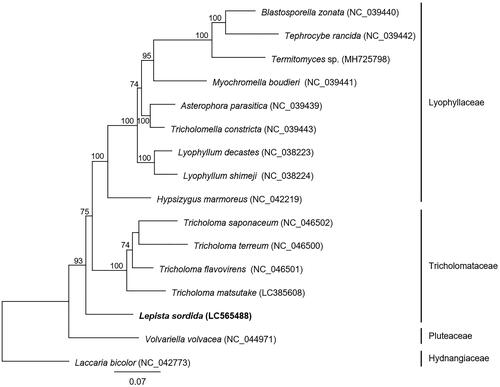
Phylogenetic analysis based on multilocus sequence analysis (MLSA) was performed using 14 conserved proteins (cox1–3, cob, nad1–6, nad4L, atp6, atp8, and atp9) from L. sordida and 15 related basidiomycetes (). The amino acid sequences of 14 proteins were individually aligned using MAFFT v7.480 (Katoh and Standley Citation2013) with the –auto option. Next, poorly aligned regions were trimmed using trimAl v1.2 (Capella-Gutierrez et al. Citation2009) with the -automated1 option to improve subsequent phylogenetic analysis. The obtained alignments were used to generate a maximum-likelihood phylogenetic tree with IQ-TREE v1.6.12 (Nguyen et al. Citation2015) using 1,000 replicates with the ultrafast bootstrap approximation approach (UFBoot2) (Hoang et al. Citation2018) implemented in IQ-TREE. The phylogenetic tree showed that L. sordida was located on the outermost branch of the Tricholomataceae clade. In the future, it will be possible to further clarify the phylogenetic relationships of the genus Lepista by obtaining the genetic information of mushrooms belonging to this genus. This is the first report of the mitochondrial genome sequence of the genus Lepista, which will open new avenues for future studies on the evolution and classification of fairy ring-forming fungi.
Figure 1. Molecular phylogenetic analysis of 16 basidiomycetes. The phylogenetic tree was constructed using the maximum-likelihood method with the amino acid sequences of 14 conserved mitochondrial proteins (cox1–3, cob, nad1–6, nad4L, atp6, atp8, and atp9). Laccaria bicolor was used as an outgroup species in the phylogenetic analysis. The accession numbers of the mitochondrial genome sequences used in this analysis are provided next to each species name. Bootstrap values higher than 70 are shown at the nodes. The scale bar indicates the number of substitutions per site.
Ethical approval
Ethical approval does not apply to this paper because the research material is mushrooms (fungus), and not human, animals, plants, or threatened/endangered species that require ethical approval.
Author contributions
Conceived and designed the research: HD. Performed the experiments: JHC, Tomohiro S, AO, MK, YT, and Toshiyuki S. Analyzed the data: Tomohiro S, AO, and HD. Wrote the manuscript: JHC, Tomohiro S, and HD. Contributed to discussions and critically reviewed the manuscript: HK. All authors reviewed the manuscript and approved the final version to be published.
Disclosure statement
No potential competing interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI (https://www.ncbi.nlm.nih.gov) under the accession number LC565488. The associated BioProject, SRA, and BioSample accession numbers are PRJDB4143, DRR327414, and SAMD00036984, respectively.
Additional information
Funding
References
- Alvarado P, Moreno G, Vizzini A, Consiglio G, Manjon JL, Setti L. 2015. Atractosporocybe, Leucocybe and Rhizocybe: three new clitocyboid genera in the Tricholomatoid clade (Agaricales) with notes on Clitocybe and Lepista. Mycologia. 107(1):123–136.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120.
- Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 25(15):1972–1973.
- Choi JH, Abe N, Tanaka H, Fushimi K, Nishina Y, Morita A, Kiriiwa Y, Motohashi R, Hashizume D, Koshino H, et al. 2010a. Plant-growth regulator, imidazole-4-carboxamide, produced by the fairy ring forming fungus Lepista sordida. J Agric Food Chem. 58(18):9956–9959.
- Choi JH, Fushimi K, Abe N, Tanaka H, Maeda S, Morita A, Hara M, Motohashi R, Matsunaga J, Eguchi Y, et al. 2010b. Disclosure of the "fairy" of fairy-ring-forming fungus Lepista sordida. Chembiochem. 11(10):1373–1377.
- Choi JH, Ohnishi T, Yamakawa Y, Takeda S, Sekiguchi S, Maruyama W, Yamashita K, Suzuki T, Morita A, Ikka T, et al. 2014. The source of "fairy rings": 2-azahypoxanthine and its metabolite found in a novel purine metabolic pathway in plants. Angew Chem Int Ed Engl. 53(6):1552–1555.
- Crusoe MR, Alameldin HF, Awad S, Boucher E, Caldwell A, Cartwright R, Charbonneau A, Constantinides B, Edvenson G, Fay S, et al. 2015. The khmer software package: enabling efficient nucleotide sequence analysis. F1000Res. 4:900.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. 2018. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 35(2):518–522.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kawagishi H. 2018. Fairy chemicals - a candidate for a new family of plant hormones and possibility of practical use in agriculture. Biosci Biotechnol Biochem. 82(5):752–758.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Kirk P, Cannon P, Minter D, Stalpers J. 2008. Ainsworth and Bisby’s dictionary of the fungi, 10th ed. Wallingford: CAB International.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Suzuki T, Yamamoto N, Choi JH, Takano T, Sasaki Y, Terashima Y, Ito A, Dohra H, Hirai H, Nakamura Y, et al. 2016. The biosynthetic pathway of 2-azahypoxanthine in fairy-ring forming fungus. Sci Rep. 6:39087.
- Takano T, Yamamoto N, Suzuki T, Dohra H, Choi JH, Terashima Y, Yokoyama K, Kawagishi H, Yano K. 2019. Genome sequence analysis of the fairy ring-forming fungus Lepista sordida and gene candidates for interaction with plants. Sci Rep. 9(1):5888.
- Tanaka Y, Suzuki T, Kurokura T, Iigo M, Toyama F, Habu N, Dohra H, Konno N. 2017. The complete genome sequence and phylogenetic analysis of the mitochondrial DNA of the wood-decaying fungus Fomitopsis palustris. Genes Genom. 39(12):1377–1385.
- Wang SY, Guo HB, Li JJ, Li W, Wang Q, Yu XD. 2019. Evaluation of five regions as DNA barcodes for identification of Lepista species (Tricholomataceae, Basidiomycota) from China. PeerJ. 7:e7307.
