Abstract
In this paper, we first report the complete mtDNA sequence of Opsarius caudiocellatus with the main aim of providing a basis for further researches of this species. Assembly circular mitogenome was 16,534 bp long encoded 13 protein-coding genes, two ribosomal RNA genes, 22 transfer RNA genes, and one control region. The gene nucleotide composition was estimated to be 28.1% A, 25.2% T, 18.6% G, 28.1% C. A maximum-likelihood (ML) phylogenetic tree was reconstructed using the 13 concatenated mitochondrial protein-coding genes of O. caudiocellatus and other 19 species of the subfamily Chedrinae. Result of the phylogenetic analysis revealed that O. caudiocellatus was well grouped with Barilius barila. This study could enrich genetic resources and be helpful to studies on evolution and conservation genetics for O. caudiocellatus.
1. Introduction
Opsarius caudiocellatus (Chu Citation1984; Cypriniformes: Danionidae: Chedrinae) is an endemic small freshwater cyprinid fish (∼111 mm), which inhabits medium to fast flowing rivers or mountain streams with rich dissolved oxygen, medium to low slope, the substrate of gravel, cobbles and large pebbles. It is mainly distributed in the Nujiang River and Lancang River in China, and it is also distributed in India, Thailand, Vietnam, and Laos. In the past, Barilius caudiocellatus was treated as the synonym name of O. caudiocellatus. The main morphological features of O. caudiocellatus are similar to those of Barilius barila such as dorsal fin iii, 7–8; anal fin ii, 9–11; pectoral fin i, 11–12; ventral fin i, 7, anal fin origin opposite the 4th–7th branching dorsal fin. O. caudiocellatus can be distinguished from Barilius barila as follows: black spots at the base of the caudal fin for O. caudiocellatus not for B. barila, and the black area is positioned in the middle of the dorsal fin for O. caudiocellatus, while on both sides of B. barila () (Chu Citation1984). The complete mitochondrial genome is regularly used in detailed distribution surveys which are urgently required for this species, and thus could more precisely reveal the taxonomic status (Qin et al. Citation2019). Nevertheless, up to the present, the complete mitochondrial genome of only four species in the genus Opsarius (O. caudiocellatus, O. canarensis, O. pulchellus, and O. bernatziki) can be retrieved in CNBI. In this paper, we firstly report the complete mtDNA sequence of O. caudiocellatus, with the main aim of providing a basis for further research and conservation of this species.
2. Materials and methods
2.1. Sample collection and preservation
The specimens were collected by gill nets from Nujiang River in Lushui County, Yunnan Province of China (25°84′75.91″ N, 98°86′69.44″ E) in May 2021, under the permission granted by Jiangsu Agri-animal Husbandry Vocational College (NSF2021ZR14). Research involving laboratory animals followed the ARRIVE guidelines (https://arriveguidelines.org/). The dead fish were selected while the live fish were released into the river. Samples were fixed in 95% ethanol and stored at the Aquatic Science and Technology Institution Herbarium (https://www.jsahvc.edu.cn/, Voucher number ASTIH-21b1108d30, Chen Xiao Jiang, [email protected]). According to the morphological characteristics description of O. caudiocellatus by Chu (Citation1984), with tools such as vernier calipers and an anatomical microscope, we identified the species by morphometry and meristematic counts.
2.2. Mitochondrial genome sequencing
In this paper, high-throughput sequencing technology was used. Genomic DNA was extracted from a specimen muscle using the Tguide Cell/tissue genomic DNA Extraction Kit (OSR-M401) (Tiangen, Beijing, China). The main experimental steps are as follows: (1) DNA sample quality control; (2) DNA Library Construction; (3) PCR Amplification; (4) Size Selection; (5) Library Quality Check; (6) Library Pooling and Sequencing, the amplified original library was submitted to Illumina HiSeq 4000 Sequencing platform (Illumina, CA, USA).
2.3. Assembly, annotation, and analysis
The quality check process was conducted on FastQC Version 0.11.8 (Andrews Citation2015) to obtain clean data, and the mitogenome was assembled from the clean data (2.46 GB) by MetaSPAdes 3.13.0 (Nurk et al. Citation2017) with Barilius malabaricus MN650735.1 as reference (Prabhu et al. Citation2020). The resulting circular contig consensus sequence was annotated and verified with MITOS WebServer (http://mitos.bioinf.uni-leipzig.de/index.py) (Bernt et al. Citation2013). Genome Map was drawn by OGDRAW (https://chlorobox.mpimp-golm.mpg.de/OGDraw.html) (Greiner et al. Citation2019). MEGA X was used for alignments, analyses, model calculation, and phylogeny reconstruction (Kumar et al. Citation2018). Maximum-likelihood (ML) phylogenetic tree was reconstructed using the concatenated mitochondrial protein-coding genes of O. caudiocellatus and 19 published species. Rasbora lateristriata (Kusuma and Kumazawa Citation2016), and Rasbora steineri (Chang et al. Citation2013) were used as outgroups to root the tree. The best evolutionary model obtained the lowest Bayesian information standard scores (Nei and Kumar Citation2000).
3. Results and discussion
3.1. Genomic analysis results
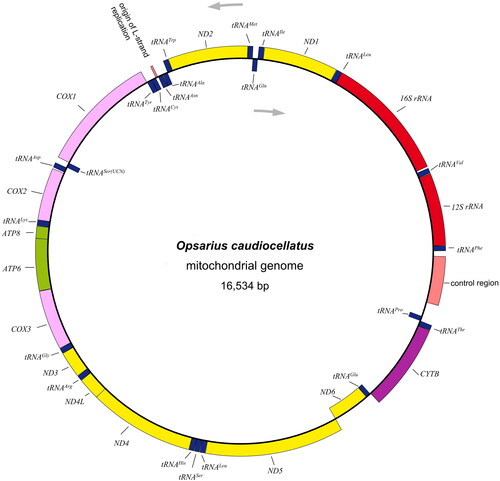
The sequenced mtDNA of O. caudiocellatus was 16,534 bp in length, comprised of typical vertebrates’ mitogenome components: 13 protein-coding genes, two ribosomal RNA genes, 22 transfer RNA genes, and one control region (CR or D-loop) (). The data was deposited at the GenBank with accession No. OM617729. Like other Chedrinae mitogenomes (Luo et al. Citation2019; Tan et al. Citation2020), nine of the thirty-seven genes (ND6, tRNAGln, tRNAAla, tRNAAsn, tRNACys, tRNATyr, tRNASer(UCN), tRNAGlu, and tRNAPro) were observed to be encoded on the L-strand (Prabhu et al. Citation2020; Chen et al. Citation2022). The overall nucleotides composition for A, T, G, and C was estimated to be 28.1%, 25.2%, 18.6%, and 28.1%, reflecting the relatively higher percentage of AT (53.3%) than GC (46.7%). CO1 and ND4L set the non-canonical GTG as the start codon as the other protein-coding genes initiated with the same ATG. Four types of stop codon usage were found, including typical termination codon TAA (ND1, CO1, ATP8, ATP6, CO3, ND3, ND4L, and ND5), TAG (ND2 and ND6), and incomplete termination codon T (CO2 and CYTB), TA (ND4). As for the protein-coding genes, the longest was ND5 (1812 bp) while the shortest was ATP8 (165 bp). Similar to most vertebrate mitogenomes, four pairs of protein-coding genes, namely ATP8-ATP6, ATP6-CO3, ND4L-ND4, and ND5-ND6, showed the overlapping which spanned 7, 1, 7,4, respectively (Wang et al. Citation2019; Yang et al. Citation2020). 22 tRNAs ranged from 65 bp of cysteine to 76 bp of lysine in length. 12S rRNA (953 bp) and 16S rRNA (1,658 bp) were the two subunits of rRNA, which were positioned between tRNAPhe and tRNALeu(UUR). The CR spanned 361 bp length and was commonly located between tRNAPro and tRNAPhe.
3.2. Phylogenetic analysis
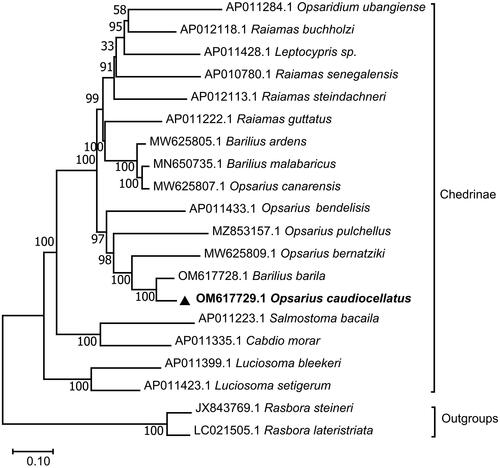
The phylogenetic position of O. caudiocellatus in subfamily Chedrinae was shown in . The phylogeny demonstrated that O. caudiocellatus clustered together with Barilius barila, Opsarius bernatziki (Yu et al. Citation2022), Opsarius pulchellus (Chen et al. Citation2022), and Opsarius bendelisis (Tang et al. Citation2010) to form a sister group of the other clade clustered by Opsaridium ubangiense (Miya et al. Citation2016), Raiamas buchholzi (Miya et al. Citation2016), Leptocypris sp. (Tang et al. Citation2010), Raiamas senegalensis (Saitoh et al. Citation2011), Raiamas steindachneri (Miya et al. Citation2016), Raiamas guttatus (Miya et al. Citation2016), Barilius ardens, Barilius malabaricus (Prabhu et al. Citation2020), and Opsarius canarensis. Further, O. caudiocellatus was closely genetically related to Barilius barila. Howes (Citation1980) divided Barilius (sensu lato) into Barilius and Opsarius (Howes Citation1980). Based on morphology (Tang et al. Citation2010) and molecular phylogeny (Liao et al. Citation2011), Qin et al. proposed that the genus Barilius from South Asia should be changed to Opsarius (Qin et al. Citation2019), so the Barilius canarensis be changed to Opsarius canarensis. Phylogenetic tree in this paper shows that Barilius and Opsarius are not monophyletic, and they gather in their respective branches. It is more reasonable that Barilius barila belongs to the genus Opsarius rather than Barilius, while Opsarius canarensis belongs to the genus Barilius rather than Opsarius.
Figure 3. Maximum-likelihood (ML) phylogenetic tree was reconstructed using the concatenated mitochondrial protein-coding genes of O. caudiocellatus and other 19 fish species. Accession numbers were given next to the species’ names. The tree topology was evaluated by 1000 bootstrap replicates. Bootstrap values at the nodes correspond to the support values for ML methods.
4. Conclusions
The complete mitochondrial genome of O. caudiocellatus was sequenced and annotated via high-throughput sequencing technology. The assembly circular mitogenome was 16,534 bp long. The phylogenetic analysis results supported that O. caudiocellatus was clustered together with B. barila. It is suggested that Barilius barila belongs to Opsarius, while Opsarius canarensis belongs to Barilius. This requires more data for further analysis and confirmation. The fundamental genetic data presented here could be beneficial for further genetic and evolutionary researches on O. caudiocellatus.
Ethical approval
Experiments were performed in accordance with the requirement of the Ethics Committee for Animal Experiments of Jiangsu Agri-animal Husbandry Vocational College. These policies were enacted according to the Chinese Association for the Laboratory Animal Sciences and the Institutional Animal Care and Use Committee (IACUC) protocols.
Authors’ contributions
Xiao Jiang Chen and Lin Song make substantial contributions to the conception or design of the work, and drafting the paper, and Final approval of the version to be published, and agreement to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved; You Wen Cao and Quan Wang were involved in the acquisition, analysis and interpretation of the data; the drafting of the paper, and the final approval of the version to be published, and agreement to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The contributions are ranked in order.
Supplemental Material
Download JPEG Image (417.1 KB)Disclosure statement
The authors declare no potential conflict of interest.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the reference number OM617729. The associated ‘BioProject,’ ‘Bio-Sample’ and ‘SRA’ numbers are PRJNA808199, SAMN26036050, and SRR18066620 respectively.
Additional information
Funding
References
- Andrews S. 2015. FastQC: a quality control tool for high throughput sequence data. Available from http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Chang CH, Tsai CL, Jang-Liaw NH. 2013. Complete mitochondrial genome of the Chinese rasbora Rasbora steineri (Teleostei, Cyprinidae). Mitochondrial DNA. 24(3):183–185.
- Chen XJ, Song L, Liu WZ, Gao P. 2022. Characterization of the complete mitochondrial genome and phylogenetic analysis of Opsarius pulchellus (Cypriniformes, Danionidae, Chedrinae). Mitochondrial DNA B. 7(4):666–668.
- Chu XL. 1984. Provisional revision of the genus Barilius in China (Pisces: Cyprinidae). Zool Res. 5(1):95–102.
- Greiner S, Lehwark P, Bock R. 2019. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47(W1):W59–W64. doi:10.1093/nar/gkz238.
- Howes GJ. 1980. The anatomy, phylogeny, and classification of bariliine cyprinid fishes. Bull Br Mus (Nat Hist) Zool. 37:129–198.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Kusuma WE, Kumazawa Y. 2016. Complete mitochondrial genome sequences of two Indonesian rasboras (Rasbora aprotaenia and Rasbora lateristriata). Mitochondrial DNA A. 27(6):4222–4223.
- Liao TY, Ünlü E, Kullander SO. 2011. Western boundary of the subfamily Danioninae in Asia (Teleostei, Cyprinidae): derived from the systematic position of Barilius mesopotamicus based on molecular and morphological data. Zootaxa. 2880(1):31–40.
- Luo F, Huang J, Luo T, Yang J, Zhou H, Wen Y. 2019. Complete mitochondrial genome and phylogenetic analysis of Yunnanilus pulcherrimus (Cypriniformes, Nemacheilidae). Mitochondrial DNA B. 4(1):1269–1270.
- Miya M, Minamoto T, Yamanaka H, Oka S-I, Sato K, Yamamoto S, Sado T, Doi H. 2016. Use of a filter cartridge for filtration of water samples and extraction of environmental DNA. Jove-J Vis Exp. (117), e54741. doi:10.3791/54741.
- Nei M, Kumar S. 2000. Molecular evolution and phylogenetics. New York (NY): Oxford University Press.
- Nurk S, Meleshko D, Korobeynikov A, Pevzner PA. 2017. MetaSPAdes: a new versatile metagenomic assembler. Genome Res. 27(5):824–834.
- Prabhu VR, Singha HS, Kumar RG, Gopalakrishnan A, Nagarajan M. 2020. Characterization of the complete mitochondrial genome of Barilius malabaricus and its phylogenetic implications. Genomics. 112(3):2154–2163.
- Qin T, Maung KW, Chen XY. 2019. Opsarius putaoensis, a new species of subfamily Danioninae (Actinopterygii, Cyprinidae) from the Irrawaddy River basin in northern Myanmar. Zootaxa. 4615(3):zootaxa.4615.3.11–593.
- Saitoh K, Sado T, Doosey MH, Bart HL, Jr Inoue JG, Nishida M, Mayden RL, Miya M. 2011. Evidence from mitochondrial genomics supports the lower Mesozoic of South Asia as the time and place of basal divergence of cypriniform fishes (Actinopterygii: ostariophysi). Zool J Linn Soc. 161(3):633–662.
- Tan HY, Yang YY, Zhang M, Chen XL. 2020. The complete mitochondrial genome of Rhinogobius duospilus (Gobiidae: gobionellinae). Mitochondrial DNA B. 5(3):3406–3407.
- Tang KL, Agnew MK, Hirt MV, Sado T, Schneider LM, Freyhof J, Sulaiman Z, Swartz E, Vidthayanon C, Miya M, et al. 2010. Systematics of the subfamily danioninae (Teleostei: cypriniformes: Cyprinidae). Mol Phylogenet Evol. 57(1):189–214.
- Wang D, Dai CX, Li Q, Li Y, Liu ZZ. 2019. Complete mitochondrial genome and phylogenic analysis of Rhinogobius cliffordpopei (Perciformes, Gobiidae). Mitochondrial DNA B. 4(2):2473–2474.
- Yang CJ, Chen Y, Chen Z, He GH, Zhong ZL, Xue WB. 2020. The next-generation sequencing reveals the complete mitochondrial genome of Rhinogobius formosanus (Perciformes: Gobiidae). Mitochondrial DNA B. 5(3):2673–2674.
- Yu P, Zhou L, Yang XL, Li LL, Jin H, Li Z, Wang ZW, Li XY, Zhang XJ, Wang Y, et al. 2022. Comparative mitogenomic analyses unveil conserved and variable mitogenomic features and phylogeny of Chedrinae fish. Zool Res. 43(1):30–32.