Abstract
The genus Allogalathea belongs to the subfamily Galatheoidea of the family Galatheidae. Here, we report a mitogenome of Allogalathea elegans (Adams & White, 1848). In this study, we obtained the complete mitochondrial genome of Allogalathea elegans by sequencing, which was 16,263 bp in length. The mitogenome contained 37 genes, including the typical set of 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, and 2 Ribosomal RNA (rRNA) genes. The nucleotides A, C, G, and T distribution was 36.40%, 19.44%, 9.09%, and 35.07%, respectively. The length of the total protein-coding genes was 11,172 bp, which accounts for 68.69% of the whole mitochondrial genome. The phylogenetic result generated by IQ-Tree based on 13 PGCs showed that the infraorder Anomura is monophyletic, and the infraorder Anomura is a sister group of the infraorder Glypheidea. The discovery of the complete mitochondrial genome of A. elegans would help to conduct in-depth research on the infraorder Anomura.
1. Introduction
Allogalathea elegans (Adams & White, 1848) belongs to the genus Allogalathea in the family Galatheidae and order Decapoda, which is mainly found in the Indian Ocean and Western Pacific Ocean (Lee et al. Citation2019). Although morphologically similar to lobsters, A. elegans is distinguished from them by having a smaller fifth pereopod, an abdomen that curves behind the thorax (Baba et al. Citation2009), a thorax that is thoroughly elongate, Flattening dorsal, and carinate in ventral, with between five and nine lateral teeth, and the carapace is covered in setiferous striae (). Some scholarly studies have shown that Allogalathea has been discovered to be species complexes with high morphological similarity, but with genetically distinct species (Cabezas et al. Citation2011). Therefore, to clarify its evolutionary status at the molecular level, we determined the mitochondrial genome sequence of A. elegans and analyzed its evolutionary characteristics, which will help us improve the data at the molecular level and clarify the phylogenetic relationship and taxonomic status of A. elegans in this study.
2. Materials and methods
2.1. Sample collection and preservation
The specimen of A. elegans was obtained from Boundary Island, Lingshui City, Hainan Province, China (E110°11′46.739″N18°35′0.386″) in November 2021. The image of A. elegans was taken by Ruan Xinhe on 25 December 2021 (). This specimen is deposited in the Laboratory of Aquatic Economic Animal Germplasm Resources and breeding Engineering, South China Agricultural University, China (Xinhe Ruan, [email protected]), under voucher number CHT2150011.
2.2. DNA extraction and sequencing and phylogenetic analysis method
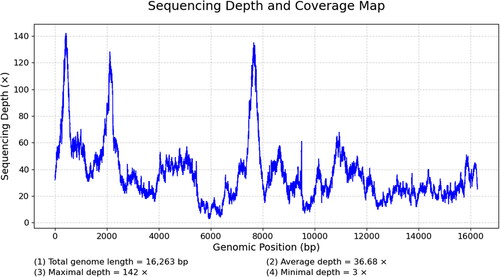
Total genomic DNA was extracted using a modified cetyltrimethylammonium bromide (CTAB) method (Allen et al. Citation2006), and NEBNext Ultra DNA Library Prep Kit for Illumina sequencing was used to construct a 500-bp paired-end library. The Illumina NovaSeq 6000 platform (BIOZERON Co., Ltd., Shanghai, China) was used for sequencing. The mitogenome was assembled from 8,051.3 Mb of raw reads with a mean depth of 36.68×. The Sequencing Depth and Coverage Map for A. elegans mitochondrial Genomes in (Ni et al. Citation2023). The obtained GC content was 44.01%. Eventually, the assembled sequence was reorganized and oriented based on the reference mitochondrial genome to generate the final assembled mitochondrial genomic sequence (Zhang et al. Citation2000; Bolger et al. Citation2014). The CPGView was used to map the mitochondrial genome (http://www.1kmpg.cn/cpgview). The base composition and phylogenetic tree were calculated using IQ-TREE 2.2.0 software (Minh et al. Citation2020). The phylogenetic tree and optimal model were constructed using IQ-TREE from nucleic acid sequences employing the maximum-likelihood method, 1000 replicates, and GTR + F+R4 model.
3. Results and discussion
3.1. Characteristics of A. elegans mitochondrial genome
The complete mitogenome of A. elegans (GenBank accession number: ON968875) was 16,263 bp long and comprised the typical set of 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, and 2 Ribosomal RNA (rRNA) genes (). All genes had similar locations and strands as those of other published squat lobsters (Lee et al. Citation2016; Zhang et al. Citation2017; Hwang et al. Citation2019). The CPGView was used to map the mitochondrial genome, as shown in . The distribution of A, C, G, and T was 36.40%, 19.44%, 9.09%, and 35.07%, respectively. The total length of the protein-coding genes was 11,172 bp, accounting for 68.69% of the mitochondrial genome, and the base composition was 28.87% A, 15.49% C, 15.47% G, and 40.18% T. Among the protein-coding genes, six genes used the start codon ATG (COX2, ATP8, ATP6, COX3, NAD4L, and COB), four genes used ATT (COX1, NAD2, NAD6, and NAD3) as the start codon, two genes used GTG (NAD5 and NAD1), and the NAD4 gene initiated with ATA codon, respectively.
Figure 3. Mitogenome pattern map of Allogalathea elegans. The mitochondrial genome is mapped using CPGView. CDS: Coding sequence; tRNA: Transfer RNA; rRNA: Ribosomal RNA.
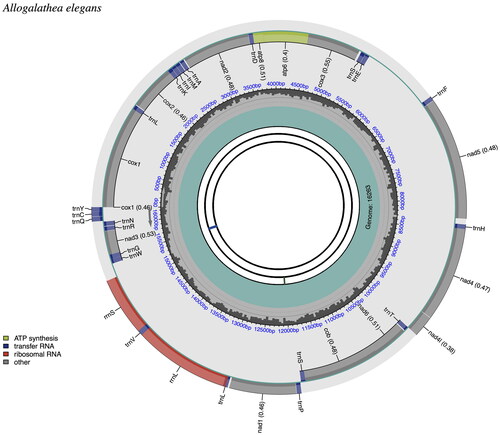
Table 1. The Organization of the complete mitochondrial genome in Allogalathea elegan.
3.2. Phylogenetic analysis
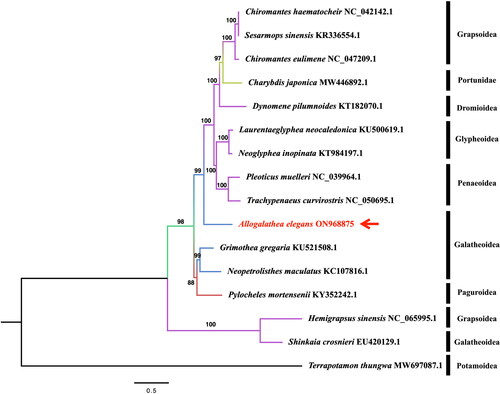
To investigate the evolutionary relationship of A. elegans, we constructed a phylogenetic tree using the maximum-likelihood (ML) method based on 13 protein-coding nucleotide sequences from the Decapoda mitogenomes. The topology and nodal support values are shown in . The results indicate that A. elegans forms a separate branch. Although A. elegans belongs to the same Galatheoidea taxonomically as Grimothea gregaria, Neopetrolisthes maculatus, and Shinkaia crosnieri, it does not belong to the same branch in the evolutionary tree, and A. elegans has a considerable genetic distance from Shinkaia crosnieri. This indicates that many of these taxa have been discovered to be species complexes demonstrating morphological similarities but with genetically distinct species (Demes et al. Citation2009). This study provides better insights into the phylogeny of this species.
Figure 4. the phylogenetic tree was based on A. elegans and other 15 species, which was performed by ML analysis of the 13 protein-coding genes. The base composition and the phylogenetic tree were calculated using IQ-TREE 2.2.0 software with the maximum-likelihood method, 1000 replicates, and GTR + F+R4 model. The phylogenetic position of A. elegans was marked with a red arrow. The following sequences were used: Chiromantes haematochir NC_042142.1 (Li et al. Citation2019), Sesarmops sinensis KR336554.1 (Xing et al. Citation2016), Chiromantes eulimene NC_047209.1 (Zhang et al. Citation2020), Charybdis japonica MW446892.1 (Liu and Cui Citation2010), Dynomene pilumnoides KT182070.1 (Shi et al. Citation2016), Laurentaeglyphea neocaledonica KU500619.1 (Tan, Gan, Dally, et al. Citation2018), Neoglyphea inopinata KT984197.1 (Tan, Gan, Dally, et al. Citation2018), Pleoticus muelleri NC_039964.1 (Kim et al. Citation2018), Trachypenaeus curvirostris NC_050695.1 (Zhu et al. Citation2019), Grimothea gregaria KU521508.1 (Lee et al. Citation2016), Neopetrolisthes maculatus KC107816.1 (Shen et al. Citation2013), Pylocheles mortensenii KY352242.1 (Tan, Gan, Lee, et al. Citation2018), Hemigrapsus sinensis NC_065995.1 (Jin et al. Citation2023), Shinkaia crosnieri EU420129.1 (Yang and Yang Citation2008), Terrapotamon thungwa MW697087.1 (Yundaeng et al. Citation2022).
4. Conclusions
We reported the first complete mitochondrial genome assembly and annotation of A. elegans using the next-generation sequencing technology. The circular mitogenome was 16,263 bp in length, contained 37 genes encoding 13 PCGs, 22 tRNAs and 2 rRNAs. The phylogenetic tree was inferred by a Maximum-likelihood phylogenetic tree based on the sequences of 18 species, which supported that A. elegans forms a separate branch and is inconsistent with the taxonomy. The mitochondrial genomic data of A. elegans provided in this study will help provide new insights into the future classification of A. elegans.
Ethical approval
This study was conducted with the guidelines of the Council of China and animal welfare requirements. Based on the recommendations of the Regulations for the Administration of Affairs Concerning Experimental Animals of China, the Institutional Animal Care and Use Committee of Guangdong Academy of Animal Science and Veterinary Medicine, South China Agricultural University approved all animal experiments (approval number: ACVM SCAU 2021002).
Author contributions
Conceived and designed the experiments: Huihong Zhao and Jie Yu. Performed the experiments: Huitao Cheng and Zijie Xuan. Analyzed the data and Wrote the paper: Xinhe Ruan and Zongyang Li. Final approval of the version to be published: Jie yu. All authors have read and agreed to the published version of the manuscript.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data supporting this study’s findings are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under accession no. ON968875. The associated Bio-Project, SRA, and Bio-Sample numbers are PRJNA904942, SRR22403925, and SAMN31859346 respectively.
Additional information
Funding
References
- Allen GC, Flores-Vergara MA, Krasynanski S, Kumar S, Thompson WF. 2006. A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat Protoc. 1(5):2320–2325. doi: 10.1038/nprot.2006.384.
- Baba K, Macpherson E, Lin CW, Chan TY. 2009. Crustacean Fauna of Taiwán. Squat lobsters (Chirostylidae and Galatheidae). National Taiwan Ocean University. http://scholars.ntou.edu.tw/handle/123456789/16148.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120. doi: 10.1093/bioinformatics/btu170.
- Cabezas P, Macpherson E, Machordom A. 2011. Allogalathea (Decapoda: Galatheidae): a monospecific genus of squat lobster? Zoolog J Linnean Soc. 162(2):245–270. doi: 10.1111/j.1096-3642.2010.00681.x.
- Demes KW, Graham MH, Suskiewicz TS. 2009. Phenotypic plasticity reconciles incongruous molecular and morphological taxonomies: the Giant Kelp, Macrocystis (laminariales, Phaeophyceae), is a monospecific genus. J Phycol. 45(6):1266–1269. doi: 10.1111/j.1529-8817.2009.00752.x.
- Hwang JY, Haque N, Lee DH, Kim BM, Rhee JS. 2019. Complete mitochondrial genome of the intertidal hermit crab, Pagurus similis (Crustacea, Anomura). Mitochondrial DNA Part B. 4(1):1861–1862. doi: 10.1080/23802359.2019.1613183.
- Jin X, Guo X, Chen J, Li J, Zhang S, Zheng S, Wang Y, Peng Y, Zhang K, Liu Y, et al. 2023. The complete mitochondrial genome of Hemigrapsus sinensis (Brachyura, Grapsoidea, Varunidae) and its phylogenetic position within Grapsoidea. Genes Genomics. 45(3):377–391. doi: 10.1007/s13258-022-01319-9.
- Kim NK, Alam J, Kim GR, Andriyono S, Park H, Kim HW. 2018. The complete mitochondrial genome of the Argentine red shrimp Pleoticus muelleri (Bate, 1888) (Crustacea, Decapoda, Solenoceridae). Mitochondrial DNA Part B. 3(2):1027–1028. doi: 10.1080/23802359.2018.1511845.
- Lee CW, Song JH, Min GS, Kim S. 2016. The complete mitochondrial genome of squat lobster, Munida gregaria (Anomura, Galatheoidea, Munididae). Mitochondrial DNA Part B. 1(1):204–206. doi: 10.1080/23802359.2016.1155087.
- Lee S, Lee S k, Kim S, Kim W. 2019. First records of two squat lobsters (Decapoda, Galatheidae) from Korea. Crustac. 92(6):725–737. doi: 10.1163/15685403-00003900.
- Li Q, Xu C, Wang C, Liu G. 2019. The complete mitochondrial genome of red-clawed crab Chiromantes haematochir (Sesarmidae: Grapsidae). Mitochondrial DNA Part B. 4(1):53–54. doi: 10.1080/23802359.2018.1536452.
- Liu Y, Cui Z. 2010. Complete mitochondrial genome of the Asian paddle crab Charybdis japonica (Crustacea: Decapoda: Portunidae): gene rearrangement of the marine brachyurans and phylogenetic considerations of the decapods. Mol Biol Rep. 37(5):2559–2569. doi: 10.1007/s11033-009-9773-2.
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von HA, Lanfear R. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evolut. 37(5):1530–1534. doi: 10.1093/molbev/msaa015.
- Ni Y, Li J, Zhang C, Liu C. 2023. Generating sequencing depth and coverage map for organelle genomes. ZappyLab, Inc. https://protocols.cloud/view/generating-sequencing-depth-and-coverage-map-for-o-cswxwffn.
- Shen H, Braband A, Scholtz G. 2013. Mitogenomic analysis of decapod crustacean phylogeny corroborates traditional views on their relationships. Mol Phylogenet Evol. 66(3):776–789. doi: 10.1016/j.ympev.2012.11.002.
- Shi G, Cui Z, Hui M, Liu Y, Chan TY, Song C. 2016. Unusual sequence features and gene rearrangements of primitive crabs revealed by three complete mitochondrial genomes of Dromiacea. Comp Biochem Physiol D. 20:65–73. doi: 10.1016/j.cbd.2016.07.004.
- Tan MH, Gan HM, Dally G, Horner S, Moreno PAR, Rahman S, Austin CM. 2018. More limbs on the tree: Mitogenome characterisation and systematic position of “living fossil” species Neoglyphea inopinata and Laurentaeglyphea neocaledonica (Decapoda). Invert System. 32(2):448–456. doi: 10.1071/IS17050.
- Tan MH, Gan HM, Lee YP, Linton S, Grandjean F, Bartholomei-Santos ML, Miller AD, Austin CM. 2018. ORDER within the chaos: insights into phylogenetic relationships within the Anomura (Crustacea: Decapoda) from mitochondrial sequences and gene order rearrangements. Mol Phylogenet Evol. 127:320–331. doi: 10.1016/j.ympev.2018.05.015.
- Xing Y, Ma X, Wei Y, Pan D, Liu W, Sun H. 2016. The complete mitochondrial genome of the semiterrestrial crab, Chiromantes neglectum (Eubrachyura: Grapsoidea: Sesarmidae). Mitochondrial DNA Part B. 1(1):461–463. doi: 10.1080/23802359.2016.1186509.
- Yang JS, Yang WJ. 2008. The complete mitochondrial genome sequence of the hydrothermal vent galatheid crab Shinkaia crosnieri (Crustacea: Decapoda: Anomura): a novel arrangement and incomplete tRNA suite. BMC Genomics. 9(1):257. doi: 10.1186/1471-2164-9-257.
- Yundaeng C, Naktang C, U-Thoomporn S, Narong N, Phetchawang P, Pootakham W, Promdam R, Tangphatsornruang S. 2022. The complete mitochondrial genome sequence of the karst-dwelling crab, Terrapotamon thungwa (Crustacea: Brachyura: Potamidae). Mitochondrial DNA Part B. 7(5):769–771. doi: 10.1080/23802359.2022.2070038.
- Zhang D, Zhou Y, Cheng H, Wang C. 2017. The complete mitochondrial genome of a yeti crab Kiwa tyleri Thatje, 2015 (Crustacea: Decapod: Anomura: Kiwaidae) from deep-sea hydrothermal vent. Mitochondrial DNA Part B. 2(1):141–142. doi: 10.1080/23802359.2017.1289347.
- Zhang Y, Gong L, Lu X, Jiang L, Liu B, Liu L, Lü Z, Li P, Zhang X. 2020. Gene rearrangements in the mitochondrial genome of Chiromantes eulimene (Brachyura: Sesarmidae) and phylogenetic implications for Brachyura. Int J Biol Macromol. 162:704–714. doi: 10.1016/j.ijbiomac.2020.06.196.
- Zhang Z, Schwartz S, Wagner L, Miller W. 2000. A greedy algorithm for aligning DNA sequences. J Comput Biol. 7(1–2):203–214. doi: 10.1089/10665270050081478.
- Zhu P, Luo P, Wang P, Xu Y, Zhang H, Wu H, Liao Y, Liang F, Liu L. 2019. The complete mitochondrial genome of Trachypenaeus curvirostris (Stimpson, 1860). Mitochondrial DNA B Resour. 4(2):2834–2835. doi: 10.1080/23802359.2019.1660279.