Abstract
The complete mitochondrial genome of Polypedilum henicurum was sequenced and analyzed. The whole mitogenome is 15,964 bp in length and contains 13 protein-coding genes (PCGs), 22 transfer RNAs (tRNAs), and two ribosomal RNAs (rRNAs). The overall nucleotide composition is 39.5% A, 39.0% T, 13.1% C, and 8.4% G. Most PCGs start with ATN codon excluding COX1 (TTG) and all PCGs end with TAA codon. Gene arrangement stays the same with the ancestral mitochondrial genome. Bayesian inference phylogenetic tree supports its close relationship with P. unifascium. This work enriches the library of Chironomidae mitochondrial genomes and provides a valuable resource for understanding the evolutionary history of Polypedilum.
Polypedilum Kieffer, 1913, is one of the largest genera in the family Chironomidae. Species delimitation and phylogeny within Polypedilum have always been conducted by morphology (Zhang et al. Citation2016). Comparative mitochondrial genome analysis provides valuable resources for understanding the evolutionary history of species (Li et al. Citation2022). While relevant analysis has not been conducted for Polypedilum because of the deficiency of mitochondrial genome data, so far, only P. nubifer, P. unifascium, and P. vanderplanki have been described with mitochondrial genomes (Deviatiiarov et al. Citation2017; Lei et al. Citation2021; Xiao et al. Citation2022). This study described a novel mitogenome of the species P. henicurum and provided essential DNA molecular database for further investigations.
Materials
P. henicurum was collected using light traps at Lishui, Zhejiang, China (27°45′16″ N, 119°11′15″ E) on August 2020. Ethical approval or permissions for sample collection were not required because the sample was an insect. The specimens were preserved in 75% ethanol and deposited at College of Life Sciences, Taizhou University (www.tzc.edu.cn, contact person: Xin Qi; e-mail: [email protected]) under the voucher number BSZ16.
Methods
The photograph of P. henicurum habitus was obtained with a DV500 5MP digital camera mounted on a Chongqing Optec SX680 stereomicroscope (). Total genomic DNA of the specimen was extracted from the abdomen of an adult using DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany). The purified DNA was fragmented and used to construct short-insert libraries (insert size 430 bp) using Illumina TruSeq Nano DNA Library Preparation Kit, and then sequenced on the Illumina NovaSeq 6000 platform. Clean reads were generated from raw reads by Trimmomatic ver. 0.40 (Bolger et al. Citation2014) and assembled using SPAdes ver. 3.14.1 (Bankevich et al. Citation2012). The assembly sequence was annotated using the MITOS web server (Bernt et al. Citation2013) and tRNAscan-SE ver. 2.0.5 (Lowe and Chan Citation2016). Some modifications were manually modified. The mitochondrial genome map was visualized using Proksee at https://proksee.ca.
Figure 1. The photograph of Polypedilum henicurum. Scale bar indicates 500 μm. Photo credits: C. Song.

To discuss the phylogenetic status of P. henicurum, mitochondrial genomes of 21 registered Chironomidae species, including five Chironominae species, five Diamesinae species, six Orthocladiinae species, two Prodiamesinae species, two Tanypodinae species and one Podonominae species, were downloaded from GenBank of NCBI, and Forcipomyia makanensis (Diptera: Ceratopogonidae: Forcipomyiinae) was used as an outgroup. The 13 PCG sequences of all the species were multiple aligned individually using MAFFT ver. 7.490 (Katoh et al. Citation2002), and then linked using SequenceMatrix ver. 1.7.8 (Vaidya et al. Citation2011). Prior to phylogenetic analyses, the substitution models were selected by PartitionFinder2 (Lanfear et al. Citation2017). The phylogenetic analyses were conducted using MrBayes (Ronquist and Huelsenbeck Citation2003) with substitution model GTR + I + G.
Results
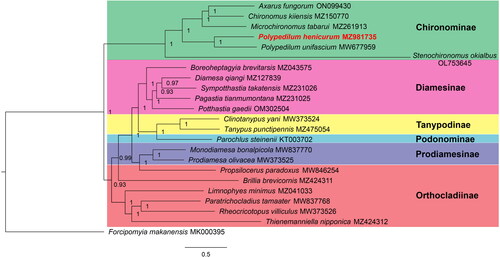
The mitochondrial genome of P. henicurum has an average depth of 294× (Figure S1). The mitogenome is a circular molecule of 15,064 bp in length, and contains 37 genes comprising 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), and two ribosomal RNA genes (rRNAs) (). The total base composition is 39.5% A, 39.0% T, 13.1% C, and 8.4% G, with an A + T bias of 78.5%. Most PCGs start with ATN codon excluding COX1 (TTG) and all PCGs end with TAA codon. Gene arrangement stays the same with the ancestral mitochondrial genome. The Bayesian inference phylogenetic tree indicated that P. henicurum belongs to the subfamily Chironominae and is close to the Polypedilum species P. unifascium ().
Figure 2. Mitochondrial genome map of Polypedilum henicurum. The CDS, tRNAs, rRNAs, GC skew and GC content are shown in the map.
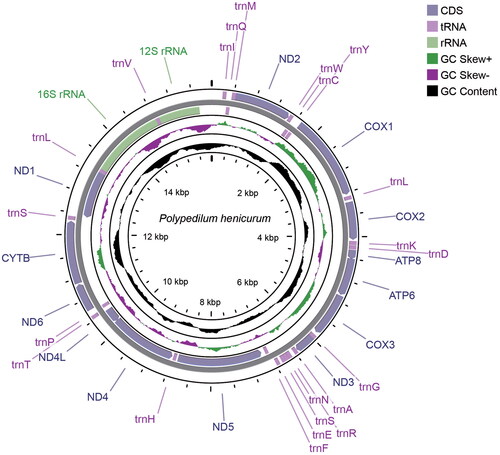
Figure 3. Bayesian inference phylogenetic tree of Chironomidae species. The tree was inferred from 22 registered Chironomidae species and one Ceratopogonidae species based on the concatenated sequences of mitogenomic PCGs. The following sequences were used: ON099430 (Qi et al. Citation2022), MZ150770 (Liu et al. Citation2022), MZ261913 (Kong et al. Citation2021), MZ981735 (this study), MW677959 (Lei et al. Citation2021), MZ043575 (Lin, Liu, et al. Citation2022), MZ127839 (Lin, Liu, et al. Citation2022), MZ231026 (Lin, Liu, et al. Citation2022), MZ231025 (Lin, Liu, et al. Citation2022), OM302504 (Lin, Liu, et al. Citation2022), MW373524 (Zheng et al. Citation2021), MZ475054 (Jiang et al. Citation2022), KT003702 (Kim et al. Citation2016), MW837770 (Lin, Zhao, et al. Citation2022), MW373525 (Zheng et al. Citation2021), MW846254 (Lin, Zhao, et al. Citation2022), MZ424311 (Lin, Zhao, et al. Citation2022), MZ041033 (Fang et al. Citation2022), MW837768 (Lin, Zhao, et al. Citation2022), MW373526 (Zheng et al. Citation2021), MZ424312 (Lin, Zhao, et al. Citation2022) and MK000395 (Jiang et al. Citation2019). Substitution model was GTR + I + G. BI posterior probability values are indicated at the nodes.
Discussion and conclusions
We described the first complete mitochondrial genome of P. henicurum. The gene content and gene arrangement were consistent with those of most registered Chironomidae mitogenomes except Stenochironomus (Zheng et al. Citation2022). The topology of Bayesian inference phylogenetic tree was supported by morphological taxonomy. These results imply the application prospect of mitogenomic data in species taxonomy. Mitogenomic sequences, gene arrangements as well as codon usage patterns are frequently used to resolve the phylogeny of organisms (Lu et al. Citation2023). To better understand the evolutionary history of Polypedilum, further studies such as codon bias analysis can be conducted based on our mitogenomic data. This work provides an essential DNA molecular database for further evolutionary and molecular research on Polypedilum. We aim to resolve the phylogenetic status of Polypedilum species with mitochondrial DNA markers, which require abundant, high-quality mitochondrial genomes from this genus in the future.
Author contributions
Xin Qi conceived the original idea and designed the work. Xuanru Di and Chao Song collected the specimens and carried out experiments. Teng Lei and Kunlun Mou analyzed experimental data. Teng Lei designed the figure and drafted the manuscript. All authors revised the final version of the manuscript and agreed to be accountable for all aspects of the work.
Supplemental Material
Download JPEG Image (971.8 KB)Disclosure statement
The authors report no conflicts of interest and are alone responsible for the content and writing of this paper.
Data availability statement
The complete mitochondrial genome of Polypedilum henicurum for this study is openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov with accession number MZ981735. The associated BioProject, BioSample, and SRA are deposited in the Genome Sequence Archive in National Genomics Data Center at https://ngdc.cncb.ac.cn under accession numbers PRJCA015227, SAMC1125797, and CRA010003.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477. doi:10.1089/cmb.2012.0021.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319. doi:10.1016/j.ympev.2012.08.023.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120. doi:10.1093/bioinformatics/btu170.
- Deviatiiarov R, Kikawada T, Gusev O. 2017. The complete mitochondrial genome of an anhydrobiotic midge Polypedilum vanderplanki (Chironomidae, Diptera). Mitochondrial DNA A DNA Mapp Seq Anal. 28(2):218–220. doi:10.3109/19401736.2015.1115849.
- Fang XL, Li XT, Lu T, Fu J, Shen M, Xiao YL, Fu Y. 2022. Complete mitochondrial genome of Limnophyes minimus (Diptera: Chironomidae). Mitochondrial DNA B Resour. 7(1):280–282. doi:10.1080/23802359.2022.2029604.
- Jiang XH, Han XJ, Liu QY, Hou XH. 2019. The mitochondrial genome of Forcipomyia makanensis (Insecta: Diptera: Ceratopogonidae). Mitochondrial DNA Part B. 4(1):344–345. doi:10.1080/23802359.2018.1544048.
- Jiang YW, Zhao YM, Lin XL. 2022. First report of the complete mitogenome of Tanypus punctipennis Meigen, 1818 (Diptera, Chironomidae) from Hebei Province, China. Mitochondrial DNA B Resour. 7(1):215–216. doi:10.1080/23802359.2021.2022544.
- Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30(14):3059–3066. doi:10.1093/nar/gkf436.
- Kim S, Kim H, Shin SC. 2016. Complete mitochondrial genome of the Antarctic midge Parochlus steinenii (Diptera: Chironomidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(5):3475–3476. doi:10.3109/19401736.2015.1066355.
- Kong FQ, Zhao YC, Chen JL, Lin XL. 2021. First report of the complete mitogenome of Microchironomus tabarui Sasa, 1987 (Diptera, Chironomidae) from Hebei Province, China. Mitochondrial DNA B Resour. 6(10):2845–2846. doi:10.1080/23802359.2021.1970638.
- Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B. 2017. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol Biol Evol. 34(3):772–773.
- Lei T, Song C, Zhu XD, Xu BY, Qi X. 2021. The complete mitochondrial genome of a non-biting midge Polypedilum unifascium (Tokunaga, 1938) (Diptera: Chironomidae). Mitochondrial DNA B Resour. 6(8):2212–2213. doi:10.1080/23802359.2021.1945977.
- Li SY, Zhao YM, Guo BX, Li CH, Sun BJ, Lin XL. 2022. Comparative analysis of mitogenomes of Chironomus (Diptera: Chironomidae). Insects. 13:1164. doi:10.3390/insects13121164.
- Lin XL, Liu Z, Yan LP, Duan X, Bu WJ, Wang XH, Zheng CG. 2022. Mitogenomes provide new insights of evolutionary history of Boreheptagyiini and Diamesini (Diptera: Chironomidae: Diamesinae). Ecol Evol. 12(5):e8957. doi:10.1002/ece3.8957.
- Lin X‐L, Zhao Y‐M, Yan L‐P, Liu W‐B, Bu W‐J, Wang X‐H, Zheng C‐G. 2022. Mitogenomes provide new insights into the evolutionary history of Prodiamesinae (Diptera: Chironomidae). Zool Scripta. 51(1):119–132. doi:10.1111/zsc.12516.
- Liu C, Xu GN, Lei T, Qi X. 2022. The complete mitochondrial genome of a tropical midge Chironomus kiiensis Tokunaga, 1936 (Diptera: Chironomidae). Mitochondrial DNA B Resour. 7(1):211–212. doi:10.1080/23802359.2021.2018952.
- Lowe TM, Chan PP. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44(W1):W54–W57. doi:10.1093/nar/gkw413.
- Lu CC, Huang XL, Deng J. 2023. Mitochondrial genomes of soft scales (Hemiptera: Coccidae): features, structures and significance. BMC Genomics. 24(1):37. doi:10.1186/s12864-023-09131-9.
- Qi Y, Duan X, Jiao KL, Lin XL. 2022. First complete mitogenome of Axarus fungorum (Albu, 1980) from Guizhou Province, China (Diptera, Chironomidae). Mitochondrial DNA B Resour. 7(10):1807–1809. doi:10.1080/23802359.2022.2131369.
- Ronquist F, Huelsenbeck JP. 2003. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19(12):1572–1574. doi:10.1093/bioinformatics/btg180.
- Vaidya G, Lohman DJ, Meier R. 2011. SequenceMatrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics. 27(2):171–180. doi:10.1111/j.1096-0031.2010.00329.x.
- Xiao YL, Xu ZG, Wang JX, Fang XL, Fu Y. 2022. Complete mitochondrial genome of a Eurytopic midge, Polypedilum nubifer (Diptera: Chironomidae). Mitochondrial DNA B Resour. 7(11):1936–1938. doi:10.1080/23802359.2022.2122746.
- Zhang RL, Song C, Qi X, Wang XH. 2016. Taxonomic review on the subgenus Tripodura Townes (Diptera: Chironomidae: Polypedilum) from China with eleven new species and a supplementary world checklist. Zootaxa. 4136(1):1–53. doi:10.11646/zootaxa.4136.1.1.
- Zheng CG, Liu Z, Zhao YM, Wang Y, Bu WJ, Wang XH, Lin XL. 2022. First report on mitochondrial gene rearrangement in non-biting midges, revealing a synapomorphy in Stenochironomus Kieffer (Diptera: Chironomidae). Insects. 13:115. doi:10.3390/insects13020115.
- Zheng CG, Zhu XX, Yan LP, Yao Y, Bu WJ, Wang XH, Lin XL. 2021. First complete mitogenomes of Diamesinae, Orthocladiinae, Prodiamesinae, Tanypodinae (Diptera: Chironomidae) and their implication in phylogenetics. PeerJ. 9:e11294. doi:10.7717/peerj.11294.
