Abstract
In the present study, we have sequenced, annotated and characterized the complete mitogenome of Tanichthys flavianalis Li, Liao & Shen, 2022, which is an endemic minnow to Hainan Province, China. The entire mitogenome size is 16,545 bp, containing 37 genes coding for 13 mitochondrial proteins, 22 tRNAs, 2 rRNAs, and one putative control region. It has the typical fish mitochondrial gene arrangement, though with 9 gene overlaps and 17 intergenic spacers scattering throughout the mitogenome, and a high A + T bias (60.8%) comparing to other Cyprinids. ATG is used as the start codon in most mitochondrial protein coding genes (PCGs) except for cox1 which uses GTG, whereas the canonical TAA/TAG are prevailing stop codons except for cox2, cox3, nad4 and cytb. Phylogenetic analyses with other reported species from Tanichyidae were conducted using IQ-TREE and MrBayes, based on the concatenated 13 PCGs. The results clustered T. flavinalis with the newly described T. albiventris Li Bohlen & Liao, 2022, and proved the reciprocally monophyletic relationship between Chinese and Vietnamese Tanichthys.
Introduction
Tanichthys is a genus of freshwater minnows in the family Tanichthyidae of Cypriniformes (Chen and Mayden Citation2009; Mayden and Chen Citation2010). They inhabit streams in southeastern China, and northern and central Vietnam. Despite being famous ornamental fish due to their bright colors (Weitzman and Chan Citation1966), until recently, the type species, T. albonubes Lin Citation1932, was the only one known. After 2000, eleven new Tanichthys species were discovered (Nguyễn and Ngô Citation2001; Bohlen et al. Citation2019; Freyhof and Herder Citation2021; Jin et al. Citation2022; Li et al. Citation2022), and so far, six of them are widely recognized (Froese and Pauly Citation2023). However, there is very little information available on these species, such as population trends and threats, habitat and ecology, and genetic resources. The insufficient information emphasizes the necessity for increased attention to this group within the academic community (Lin Citation1932; Nguyễn and Ngô Citation2001; Freyhof and Herder 2011; Bohlen et al. Citation2019; Li et al. Citation2022; Yang et al. Citation2023).
Since 1989, wild populations of Tanichthys have held a class II status under state protection in China (Yue and Chen Citation1998). Recent studies have identified multiple cryptic species within the genus, shedding light on the potentially extensive genetic diversity of Tanichthys (Jin et al. Citation2022; Li et al. Citation2022). Over the years, Tanichthys’ taxonomic classification has been a topic of contention, with debates on its placement within various subfamilies, such as Leuciscinae (Chu Citation1935; Yang and Huang Citation1964), Danioninae (Chen Citation1998; Nelson et al. Citation2016), Tincinae (Tang et al. Citation2010), or as a sister group to Acheilognathinae within the family Cyprinidae (Liao et al. Citation2011). However, the prevailing consensus now leans toward establishing a new family, Tanichthyidae, to accommodate Tanichthys. This shift in classification aligns with the elevation of the former Cyprinidae to superfamily status, supported by emerging molecular phylogenetic evidence (Chen and Mayden Citation2009; Mayden and Chen Citation2010). Within this updated taxonomic framework, our focal species, T. flavianalis, takes its place as a recently discovered member of the Tanichthys genus, originating from Hainan Province, China. It sets itself apart from its congeners with a higher number of branched dorsal- and anal-fin rays, and distinctive reddish-orange and golden margins on the dorsal- and anal-fins, respectively (). Studying the mitochondrial genome of T. flavianalis holds significance as it provides further genetic insights into this species, aiding in a deeper understanding of the genetic diversity and evolutionary history of Tanichthys, while also serving as valuable scientific groundwork for wildlife conservation and protection efforts.
Materials
A T. flavianalis was collected from the upper reach of the Jiuqu River (110.2861°E, 19.0451°N) in Qionghai City, Hainan Province, China, 15 June 2020, and euthanized with eugenol, then preserved in 95% ethanol, and stored in the Aquatic Museum of Xinyang Agricultural and Forestry University (https://www.xyafu.edu.cn/, Liangjie Zhao, [email protected], voucher number XYAFU-Mu-1803245).
Methods
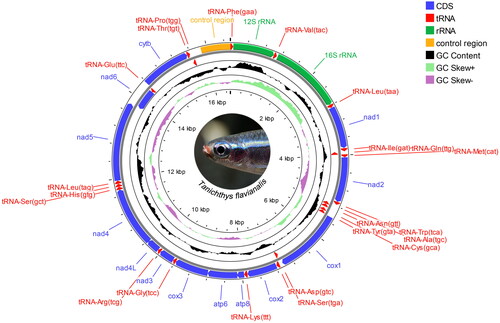
Genomic DNA was extracted from caudal-fin using the Ezup Column Animal Genomic DNA Purification Kit (#B518251-0100, Sangon Biotech, Shanghai, China) and verified for quality and quantity using 1.2% agarose gel electrophoresis and NanoDrop2000 Spectrophotometers (Thermo Scientific, USA), respectively. Isolated DNA was stored at −20 °C, until we used long and accurate PCR (LA-PCR) with nine pairs of primers designed specifically for Tanichthys species (Table S1) to amplify the mitochondrial sequences. The LA-PCR Kit (#B639285-0100, Sangon Biotech) was used for 50 μL volume reactions which contain 1.5 μL template DNA (2 ng/μL), 5 μL 10 × Long PCR Buffer, 1 μL dNTP Mix (10 mM), 0.5 μL each primer (10 μM), 0.5 μL Long and Accurate Enzyme, and 41 μL sterilized ddH2O. PCR conditions were adjusted based on primer TM values and amplicon sizes according to the manufacturer’s instruction. PCR products were sequenced using an ABI 3730XL DNA analyzer (Applied Biosystems, Foster City, CA). The sequencing result was uploaded to GenBank as a fastq file containing base-calling quality information. Amplicons ranged from 850 bp to 3876 bp (see Figure S1, supplementary material), empowering us to assemble the entire mitogenome of the species by using DNAMAN (ver8.0.8, Lynnon Biosoft, Canada, see Figure S2, supplementary material). MITOS2 WebServer (Donath et al. Citation2019) was used to annotate the assembled mitogenome, and the circular display was made by using CGViewBuilder in Proksee web server (Stothard and Wishart Citation2005) ().
Figure 2. Circular map of the assembled Tanichthys flavianalis mitogenome. Gene encoded on H- and L- strands with inverse arrow directions are shown outside and inside the circle, respectively. Plots of GC content and skew used a window size of 500 and reflect GC content/skew on a scale of 0 to 1 using a baseline of 0.5. Positive and negative skew are indicated by values above and below the midpoint respectively. This map was drawn using the Proksee web server.
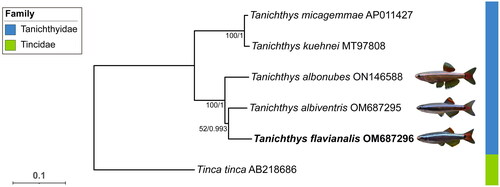
In order to understand the phylogenetic position of T. flavianalis within the Tanichthys genus, we collected all reported mitochondrial reference genomes of species in Tanichthyidae from GenBank. Tinca tinca was introduced as an outgroup. The corresponding sequence accession numbers were listed in the legend of 13 PCGs of each mitogenome were extracted, and then aligned. The alignment was performed using ‘L-INS-I’ strategy in MAFFT v7.505 (Katoh and Standley Citation2013). We concatenated 13 aligned mitochondrial PCGs blocks, each contains six sequences, into a multi-locus dataset as the input for downstream phylogenetic analyses. Maximum-likelihood (ML) phylogenies were inferred using IQ-TREE v2.2.0 (Nguyen et al. Citation2015) under Edge-linked partition model with 1000 standard bootstrap, and Bayesian inference (BI) using MrBayes v3.2.7a (Ronquist et al. Citation2012) ran four simultaneous MCMC chains for one million generations with every 1000 generations to sample. The first 25% of samples were discarded as burn-in. ModelFinder v1.6.12 (Kalyaanamoorthy et al. Citation2017) embedded in PhyloSuite v1.2.3 (Xiang et al. Citation2023) was employed to determine the optimal partitioning strategies and the corresponding models. Visualization of the resulted trees was achieved by using iTOL (Letunic and Bork Citation2019).
Figure 3. The phylogenetic tree constructed based on 13 protein coding genes of five Tanichthyidae mitogenomes and an outgroup. The support values of the corresponding nodes are shown (bootstrap values for ML/Bayes posterior probabilities for BI). The focal species is in bold, and pictures of the three Chinese Tanichthys were provided by Fan Li. The following sequences were used: T. micagemmae AP011427 (unpublished), T. kuehnei MT97808 (unpublished), T. albonubes ON146588 (unpublished), T. albiventris OM 687295 (Yang et al. Citation2023), T. flavianalis OM687296 (this study), and Tinca tinca AB218686 (Saitoh et al. Citation2006).
Results
T.flavianalis has a 16545 bp-long mitogenome, and it consists of 13 PCGs, 22 tRNA genes, 2 rRNA genes, and a noncoding AT-rich control region (). There are nine gene overlaps (from 1 to 7 bp in size) and 17 intergenic spacers (from 1 to 199 bp) in the mitogenome. The most overlapped fragment is between atp8 and atp6, while the longest spacer locates between tRNA-Pro and tRNA-Phe, which contains the control region. Of the 13 PCGs, codon ATG is overwhelmingly used as the start codon with the exception of cox1, which starts with GTG. All PCGs terminate with canonical stop codon TAA or TAG except for four genes that have incomplete stop codons of ‘T-’ (cox2 and cytb) or ‘TA-’ (cox3 and nad4). The overall nucleotide composition of the T. flavianalis mitogenome is 31.34% A (5185), 29.47% T (4876), 15.50% G (2565), and 23.69% C (3919).
Topological consistency is observed between the ML and BI trees of Tanichthyidae species. Here we display the ML tree and indicate the branch support values with both the standard bootstrap value, and the Bayes posterior probability (). The two clades within Tanichthys, which could be told apart by their geographical distributions (T. micagemmae and T. kuehnei from Vietnam, and T. albonubes, T. flavianalis and T. albiventris from China), were in reciprocal monophyly.
Discussion and conclusion
In the mitogenome of T. flavianalis, all PCGs are encoded in the heavy strand (H) except nad6, fourteen tRNA genes are located in the H-strand and the remaining eight tRNAs are in L-strand, which is consistent with most fish mitogenomes (Billington and Hebert Citation1991; Satoh et al. Citation2016). T. flavianalis shows a relatively high A + T bias (60.81%) in its mitogenome when compared to other Cyprinidae species (Tang et al. Citation2010; Liaw et al. Citation2013; Luo et al. Citation2019; Zhang et al. Citation2021), but coincides with the situation in other Tanichthys species (Yang et al. Citation2023).
Our mitogenome-based phylogenetic analyses support the placement of T. flavianalis to the genus Tanichthys, and show its close relationship to other Chinese Tanichthys species, which is consistent with previous morphologically based conclusion (Li et al. Citation2022). Our observation of phylogenetic connections aligning with geographical regions implies that local environmental factors may have driven speciation in this genus. This situation, in particular, raise intriguing questions about the possible presence of undescribed cryptic species within widely distributed species, such as T. albonubes. Further investigation into the genetic diversity of Tanichthys has the potential to enhance our understanding of the evolutionary history and conservation considerations for these fascinating fish species.
Ethical approval
Experiment permission was issued by the Ethics Committee of Xinyang Agriculture and Forestry University (XYNLXY20210911), and all procedures involving animal attendances were strictly followed the Chinese Association for the Laboratory Animal Sciences and the Institutional Animal Care and Use Committee (IACUC) protocols.
Author contributions
LZ and FL conceived and designed the study, and conducted fundraising. FL and CS collected the samples. TY performed the experiments and uploaded the sequence information to GenBank. YG analyzed data and wrote the manuscript. All authors have proofread the manuscript and agreed to publish this version.
Supplemental Material
Download JPEG Image (54.7 KB)Supplemental Material
Download JPEG Image (110.9 KB)Supplemental Material
Download MS Word (13.2 KB)Supplemental Material
Download PDF (344 KB)Supplemental Material
Download MS Word (14.7 KB)Acknowledgements
We thank four anonymous reviewers for their valuable comments on this article.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data of the study are deposited in GenBank of NCBI (https://www.ncbi.nlm.nih.gov/) under the accession no. OM687296. The associated BioProject, Bio-Sample and Run numbers are PRJNA957284, SAMN34248125, and SRR25113585, respectively.
Additional information
Funding
References
- Billington N, Hebert PD. 1991. Mitochondrial DNA diversity in fishes and its implications for introductions. Can J Fish Aquat Sci. 48(S1):80–94. doi: 10.1139/f91-306.
- Bohlen J, Dvořák T, Thang HN, Šlechtová V. 2019. Tanichthys kuehnei, new species, from Central Vietnam (Cypriniformes: Cyprinidae). Ichthyol Explor Freshw. 1081:1–10.
- Chen YY. 1998. Fauna Sinica, Osteichthyes, Cypriniformes II. Beijing: Science Press. (in Chinese with English abstract)
- Chen WJ, Mayden RL. 2009. Molecular systematics of the Cyprinoidea (Teleostei: Cypriniformes), the world’s largest clade of freshwater fishes: further evidence from six nuclear genes. Mol Phylogenet Evol. 52(2):544–549. doi: 10.1016/j.ympev.2009.01.006.
- Chu YT. 1935. Comparative studies on the scales and on the pharyngeals and their teeth in Chinese cyprinids, with particular reference to taxonomy and evolution. Biol Bull St. John’s Univ Shanghai. 2:1–225.
- Donath A, Jühling F, Al-Arab M, Bernhart SH, Reinhardt F, Stadler PF, Middendorf M, Bernt M. 2019. Improved annotation of protein-coding genes boundaries in metazoan mitochondrial genomes. Nucleic Acids Res. 47(20):10543–10552. doi: 10.1093/nar/gkz833.
- Freyhof J, Herder F. 2021. Tanichthys micagemmae, a new miniature cyprinid fish from Central Vietnam (Cypriniformes: Cyprinidae). Ichthyol Explor Freshw. 12:215–220.
- Froese R, Pauly D. Editors. 2023. FishBase. World Wide Web electronic publication. www.fishbase.org, version (02/2023).
- Jin J, Li C, Zhao J. 2022. Descriptions of six new species in White Cloud Mountain minnow Tanichthys albonubes complex (Cypriniformes: Tanichthyidae) in southern China. J Fish Biol. 100(4):1062–1087. doi: 10.1111/jfb.15012.
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589. doi: 10.1038/nmeth.4285.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. doi: 10.1093/molbev/mst010.
- Letunic I, Bork P. 2019. Interactive tree of life (itol) v4: recent updates and new developments. Nucleic Acids Res. 47(W1):W256–W259. doi: 10.1093/nar/gkz239.
- Li F, Liao T-Y, Bohlen J, Shen Z-X, Zhao L-J, Li S. 2022. Two new species of Tanichthys (Teleostei: Cypriniformes) from China. J Verteb Biol. 71(21067):1–13.
- Liao TY, Ünlü E, Kullander SO. 2011. Western boundary of the subfamily Danioninae in Asia (Teleostei, Cyprinidae): derived from the systematic position of Barilius mesopotamicus based on molecular and morphological data. Zootaxa. 2880(1):31–40. doi: 10.11646/zootaxa.2880.1.3.
- Liaw NHJ, Tsai CL, Watanabe K. 2013. Complete mitochondrial genome of the Kikuchi’s minnow Aphyocypris kikuchii (Teleostei, Cyprinidae). Mitochondiral DNA. 24(1):11–13. doi: 10.3109/19401736.2012.710227.
- Lin S-Y. 1932. New cyprinid fishes from white cloud mountain, Canton. Lingnan Sci J. 11:379–383.
- Luo F, Luo T, Huang J, Liu X, Ling S, Wen Y. 2019. Characterization of complete mitochondrial genome of Aphyocypris pulchrilineata (Teleostei, Cypriniformes, Cyprinidae). Mitochondrial DNA B. 4(1):1267–1268. doi: 10.1080/23802359.2019.1591221.
- Mayden RL, Chen WJ. 2010. The world’s smallest vertebrate species of the genus Paedocypris: a new family of freshwater fishes and the sister group to the world’s most diverse clade of freshwater fishes (Teleostei: Cypriniformes). Mol Phylogenet Evol. 57(1):152–175. doi: 10.1016/j.ympev.2010.04.008.
- Nelson JS, Grande TC, Wilson MVH. 2016. Fishes of the world, 5th ed. New Jersey: John Wiley & Sons.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. doi: 10.1093/molbev/msu300.
- Nguyễn VH, Ngô SV. 2001. Freshwater fishes of Vietnam, vol. 1. Hanoi, Vietnam: Family Cyprinidae. Aquaculture Publishing House. (in Vietnamese)
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542. doi: 10.1093/sysbio/sys029.
- Saitoh K, Sado T, Mayden RL, Hanzawa N, Nakamura K, Nishida M, Miya M. 2006. Mitogenomic evolution and interrelationships of the Cypriniformes (Actinopterygii: Ostariophysi): the first evidence toward resolution of higher-level relationships of the world’s largest freshwater fish clade based on 59 whole mitogenome sequences. J Mol Evol. 63(6):826–841. doi: 10.1007/s00239-005-0293-y.
- Satoh TP, Miya M, Mabuchi K, Nishida N. 2016. Structure and variation of the mitochondrial genome of fishes. BMC Genomics. 17(1):719. doi: 10.1186/s12864-016-3054-y.
- Stothard P, Wishart DS. 2005. Circular genome visualization and exploration using CGView. Bioinformatics. 21(4):537–539. doi: 10.1093/bioinformatics/bti054.
- Tang KL, Agnew MK, Vincent Hirt M, Sado T, Schneider LM, Freyhof J, Sulaiman Z, Swartz E, Vidthayanon C, Miya M, et al. 2010. Systematics of the subfamily Danioninae (Teleostei: Cypriniformes: Cyprinidae). Mol Phylogenet Evol. 57(1):189–214. doi: 10.1016/j.ympev.2010.05.021.
- Weitzman SH, Chan LL. 1966. Identification and relationships of Tanichthys albonubes and Aphyocypris pooni, two cyprinid fishes from South China and Hong Kong. Copeia. 1966(2):285–296. doi: 10.2307/1441136.
- Xiang C-Y, Gao F, Jakovlić I, Lei H-P, Hu Y, Zhang H, Zou H, Wang G-T, Zhang D. 2023. Using PhyloSuite for molecular phylogeny and tree-based analyses. iMeta. 2(1):e87. doi: 10.1002/imt2.87.
- Yang GR, Huang JH. 1964. Leuciscinae. In: Wu HW, Yang GR, Huang HJ, editors. Cyprinid fishes of China, vol 1. Shanghai: shanghai Science Technology Press. (in Chinese)
- Yang T, Tan C, Sun W, Li D, Zhao L, Li F. 2023. Characterization of the complete mitochondrial genome sequence of Tanichthys albiventris and phylogenetic analysis. Mitochondrial DNA B Resour. 8(2):207–210. doi: 10.1080/23802359.2023.2171244.
- Yue PQ, Chen YY. 1998. China red data book of endangered animals: Pisces. Beijing: science Press.
- Zhang Z, Li S, Zhang J, Song W, Yang J, Mu J. 2021. The complete mitochondrial genome of an endangered minnow Aphyocypris lini (Cypriniformes: Xenocyprididae): genome characterization and phylogenetic consideration. Biologia. 76(11):3311–3321. doi: 10.1007/s11756-021-00811-z.

