Abstract
Pulsatilla chinensis f. alba D. K. Zang 1993 is a forma of Pulsatilla chinensis (Bge.) Regel, the root of P. chinensis is traditional Chinese medicine called Pulsatillae radix. The biggest difference between P. chinensis f. alba and P. chinensis is that P. chinensis f. alba sepals is white. The complete chloroplast genome of P. chinensis f. alba was sequenced using the Illumina NovaSeq platform for the first time. The lengths of the genome, large single-copy (LSC), small single-copy (SSC), two inverted repeats (IRs), and GC content were 163,654 bp, 82,355 bp, 19,069 bp, 31,115 bp, and 37.2%, respectively. It had 134 genes, including 90 protein-coding genes, 36 tRNA genes, and eight rRNA genes. The maximum-likelihood tree indicated that P. chinensis f. alba had a closer relationship with P. chinensis. This study would provide a theoretical basis for the further study of Pulsatilla plants genetics phylogenetic research.
Introduction
Pulsatilla chinensis f. alba D. K. Zang 1993, a perennial herb of Ranunculaceae, is first discovered in Shandong Province, China, which is the forma of Pulsatilla chinensis (Bunge) Regel 1861 (Liang et al. Citation1993). It was also observed in Liaoning Province of China (). P. chinensis f. alba is different from P. chinensis in the color of their sepals. The sepals of P. chinensis f. alba exhibited a white coloration, while the sepals of P. chinensis displayed a violet hue. The dry root of P. chinensis, named Pulsatillae Radix, is traditional Chinese medicine included in the Chinese Pharmacopeia (National Pharmacopoeia Commissions Citation2020). Modern pharmacological research demonstrates that Pulsatilla has the traditional antipyretic and antidysentery, anti-microorganism, anti-sucrose enzyme, and antitumor effect (Li and Lin Citation2005). As a forma of P. chinensis, P. chinensis f. alba is an excellent ornamental flower and an important germplasm resource for cultivating new varieties of P. chinensis. In this study, the complete chloroplast genome of P. chinensis f. alba was sequenced and reported for the first time. It would provide research data for the evolutionary relationships of Pulsatilla plants and the cultivation and breeding of P. chinensis.
Figure 1. Pulsatilla chinensis f. alba D. K. Zang. Leaves 4 or 5, not fully expanded at anthesis; densely long pilose; leaf blade broadly ovate; 3-foliolate; margin entire or toothed. Sepals white. Wildlife photos were taken by C.B. in the Chinese city of Liaoyang, Liaoning Province. (E 123°33′09.26″, N 41°42′16.12″).

Materials and methods
Plant material and DNA sequence
We have obtained permission from the local forestry department to collect samples. This study was conducted by the laws of the People’s Republic of China. Fresh leaves of P. chinensis f. alba was collected from Liaoyang, China (E 123°33′09.26″, N 41°42′16.12″), and identified by professor Liang Xu in Liaoning University of Traditional Chinese Medicine. The voucher specimen and genomic DNA were deposited at the herbarium of Liaoning University of Chinese Medicine (Liang Xu [email protected], P. chinensis f. alba number: 10162230517007LY) (Supplemental Figure S1). The genomic DNA was stored in the Key Laboratory of Traditional Chinese Medicine at the University (Dalian, China) (Liang Xu [email protected]).
Total genomic DNA was extracted from 150 mg fresh leaves using the cetyltrimethylammonium bromide method (Doyle and Doyle Citation1987). An aliquot of purified DNA (1 μg) was then fragmented to construct a short-insert (350 bp) library using the Nextera XT DNA library preparation kit (Illumina, San Diego, CA). The library was sequenced using the Illumina NovaSeq 6000 platform, and the coverage was measured using samtools depth.
Genome assembly and annotation
The raw data was edited using the NGS QC Tool Kit v2.3.3 (https://nipgr.ac.in/ngsqctoolkit.html) (Patel and Jain Citation2015). High-quality sequence data (4.09 G) were then selected for the de novo assembly of the complete chloroplast genome using the assembler SPAdes v3.14.1 (http://cab.spbu.ru/software/spades/) (Bankevich et al. Citation2012). Finally, the complete chloroplast genome was annotated using PGA (Qu et al. Citation2019) with the chloroplast genome of Pulsatilla chinensis var. kissii (MK860683) as a reference. The maps of the chloroplast genome, cis-splicing genes, and trans-splicing genes of P. chinensis f. alba were processed by CPGview (Liu et al. Citation2023). The mVista (http://genome.lbl.gov/vista/mvista/submit.shtml) was used to analyze the similarities of P. chinensis f. alba and four other published P. chinensis chloroplast genomes (NC039452, MK860682, MK569491, MK860683) in Shuffle-LAGAN mode with P. chinensis (MK860682) as a reference.
Phylogenetic analysis
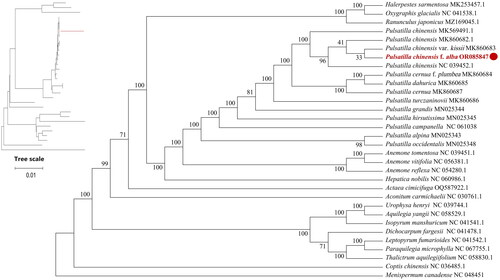
Phylogenetic tree can be represented by branching diagrams, which illustrate the relationships between similar organisms in a tree-like structure (Feng Citation2009). To analyze the phylogenetic relationship of Pulsatilla genus, 30 other complete chloroplast genomes from Ranunculaceae plants (14 from the Pulsatilla genus), one outgroup taxon from Menispermaceae (Menispermum canadense), were obtained from NCBI. The MAFFT version 7.037 (Katoh and Standley Citation2013) was used to identify common protein-coding genes of 32 chloroplast genomes and compare the P. chinensis f. alba chloroplast genome with 31 other complete chloroplast genomes by the FFT-NS-2 strategy. The gaps in the alignment were trimmed using Gblocks (Version 0.91b, http://molevol.cmima.csic.es). A phylogenetic tree of the 32 chloroplast genomes was then constructed using IQ-TREE-1.6.12, (http://www.iqtree.org/) based on the maximum-likelihood method with 1000 bootstrap replications and the JTT + F+R2 model, which was selected using ModelFinder (Kalyaanamoorthy et al. Citation2017).
Results
Genome structure analysis
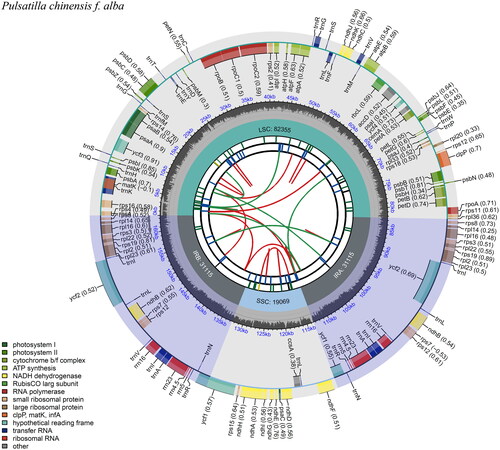
The chloroplast genome of P. chinensis f. alba was 163,654 bp, including a large single-copy (LSC) region with a length of 82, 355 bp, a small single-copy (SSC) region with a length of 19,069 bp, and two inverted repeat regions (IRs) with a length of 31,115 bp (). The genome had 134 genes, including 90 protein-coding genes, 36 tRNA genes, and eight rRNA genes, with a GC content of 37.2%. The rps16, trnK-UUU, rpoC1, atpF, trnG-UCC, trnL-UAA, trnV-UAC, petB, petD, rpl16, rpl2, ndhB, trnI-GAU, trnA-UGC and ndhA genes contained one intron; the clpP and ycf3 genes contained two introns (Figure S2). The rps12 gene was a trans-spliced gene (Figure S3). The chloroplast genomes of P. chinensis f. alba and four other published P. chinensis (NC039452, MK860682, MK569491, MK860683) had high similarity (Figure S4). The chloroplast genome of P. chinensis f. alba was correctly assembled according to the coverage depth (Average sequencing depth was 389.92X, Maximal sequencing depth was 616X, Minimal sequencing depth was 33X) (Figure S5) (Li et al. Citation2009; Li Citation2013).
Figure 2. Chloroplast genome map of Pulsatilla chinensis f. alba D. K. Zang. From the center going outward, the first circle shows the forward and reverse repeats connected with red and green arcs, respectively. The second and third circles show the tandem repeats and microsatellite sequences marked with short bars, respectively. The outer circle shows the gene structure of the chloroplast genome. The genes were colored based on their functional categories, which were shown in the left corner. The map was drawn by cpgview (Liu et al. Citation2023).
Phylogenetic analysis
Menispermum canadense was the outgroup, which was distant from the other species. P. chinensis f. alba, three P. chinensis, and P. chinensis var. kissii formed a single branch and there was a relatively short genetic distance between P. cernua and P. dahurica. The phylogenetic tree analysis revealed that Anemone is the closest relative to Pulsatilla within the Ranunculaceae ().
Figure 3. Maximum-likelihood (ML) phylogenetic tree of P. chinensis f. alba and 31 other complete chloroplast genome sequences. The numbers above the branches indicate the bootstrap values from ML analyses. The best evolutionary model was chosen as JTT + F+R2, which was selected using ModelFinder. The scale bar in the lower left corner of the figure represents the evolutionary distance, with a unit length of 0.01. The following sequences were used: Halerpestes sarmentosa (MK253457) (He et al. Citation2019), Oxygraphis glacialis NC_041538 (Zhai et al. Citation2019), Ranunculus japonicus MZ169045, P. chinensis MK569491, P. chinensis MK860682 (Zhang et al. Citation2019), Pulsatilla chinensis var. kissii MK860683 (Zhang et al. Citation2019), P. chinensis NC_039452 (Liu et al. Citation2018), P. cernua f. plumbea MK860684 (Zhang et al. Citation2019), P. dahurica MK860685 (Zhang et al. Citation2019), P. turczaninovii MK860686, P. cernua MK860687 (Zhang et al. Citation2019). P. grandis MN025344 (Li et al. Citation2020), P. hirsutissima MN025345 (Li et al. Citation2020), P. campanella NC_061038 (Xue et al. Citation2022), P. alpina MN025343 (Zhang et al. Citation2019), P. occidentalis MN025348 (Li et al. Citation2020). Anemone. tomentosa NC_039451 (Liu et al. Citation2018), A. vitifolia NC_056381, A. reflexa NC_054280 (Zhang et al. Citation2021), Hepatica nobilis NC_060986, Actaea cimicifuga OQ587922, Aconitum carmichaelii NC_030761, Urophysa henryi NC_039744, Aquilegia yangii NC_058529, Isopyrum manshuricum NC_041541 (Zhang et al. Citation2019), Dichocarpum fargesii NC_041478 (Zhang et al. Citation2019), Leptopyrum fumarioides NC_041542 (Zhang et al. Citation2019), Paraquilegia microphylla NC_067755, Thalictrum aquilegiifolium NC_058830, Coptis chinensis NC_036485, Menispermum canadense NC_048451.
Discussion and conclusion
The chloroplast genome of P. chinensis f. alba was reported in this study. The structure, size, and genetic composition of this genome were similar to those of P. chinensis and other plants in the Pulsatilla genus (Zhang et al. Citation2019). Based on the phylogenetic analysis results, P. chinensis f. alba had very close relation to P. chinensis. The above results support traditional morphological classification, and this study also provides data for the study of evolutionary relationships in the Ranunculaceae plants, the identification of Pulsatillae radix and development of germplasm resources.
Ethics statement
According to the Wild Plants Protection Regulations of the People’s Republic of China, P. chinensis f. alba was not included on the list of national protected wild plants. Article five also encourages scientific research on wild plants. Therefore, the sample collection does not require any permission. This study protocol has been approved by the School of Pharmacy, Liaoning University of Traditional Chinese Medicine. All operation was conducted in compliance with the guidelines in Specification on Good Agriculture and Collection Practices for Medicinal Plants (GACP; number: T/CCCMHPIE 2.1-2018).
Author contributions
C.B. and Y.P.X.: Analysis of data, conception and drafting for the work. Y.Y.Y. and L.X.: Identification and collection of the plant, preservation of plant specimens and conceiving the work. T.G.K.: Revising critically important intellectual content and was involved in validation and supervision. H.F.X. and W.J.H: Acquisition and analysis of data. W.J.H. and Y.P.X.: Contributed to the writing and revising. All authors agree to be accountable for all aspects of the work.
Supplemental Material
Download MS Word (1 MB)Disclosure statement
No potential conflict of interest was reported by the authors. Che Bian and Yan-ping Xing contributed equally to this research. It is worth noting that Che Bian and Yan-ping Xing are co-first authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under accession NO. OR085847. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA978389, SRX20587082 (Illumina), and SAMN35555254, respectively.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477. doi: 10.1089/cmb.2012.0021.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Feng SL. 2009. Research on method of the construction of phylogenetic trees. Inform Technol. 33(6):38–40 + 44.
- He J, Yao M, Lyu RD, Lin LL, Liu HJ, Pei LY, Yan SX, Xie L, Cheng J. 2019. Structural variation of the complete chloroplast genome and plastid phylogenomics of the genus Asteropyrum (Ranunculaceae). Sci Rep. 9(1):15285. doi: 10.1038/s41598-019-51601-2.
- Kalyaanamoorthy S, Minh BQ, Wong TK, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589. doi: 10.1038/nmeth.4285.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. doi:10.1093/molbev/mst010.
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv: Genomics. (0):3.
- Liang YT, Zang DK, Ren YG. 1993. Two new forms of Pulsatilla chinensis (Bunge) Regel from Shandong. Bull Entomol Res. 13(4):340–341.
- Li H, Handsaker B, Wysoker A, Fennel T, Ruan J, Homer N, Marth G, Abecasis GC, Durbin R. 2009. The sequence alignment/Map format and SAMtools. Bioinformatics. 25(16):2078–2079. doi: 10.1093/bioinformatics/btp352.
- Li Y, Lin CY. 2005. A summary of the chemical constituents and activities of the Pulsatilla. Tianjin J Tradition Chinese Med. 22(06):526–528.
- Li QJ, Su N, Zhang L, Tong RC, Zhang XH, Wang JR, Chang ZY, Zhao L, Potter D. 2020. Chloroplast genomes elucidate diversity, phylogeny, and taxonomy of Pulsatilla (Ranunculaceae). Sci Rep. 10(1):19781. doi: 10.1038/s41598-020-76699-7.
- Liu HJ, He J, Ding CH, Lyu R, Pei LY, Cheng J, Xie L. 2018. Comparative analysis of complete chloroplast genomes of Anemoclema, Anemone, Pulsatilla, and Hepatica revealing structural variations among Genera in Tribe Anemoneae (Ranunculaceae). Front Plant Sci. 9:1097. doi: 10.3389/fpls.2018.01097.
- Liu S, Ni Y, Li J, Zhang X, Yang H, Chen H, Liu C. 2023. CPGView: a package for visualizing detailed chloroplast genome structures. Mol Ecol Resour. 0:1–11.
- National Pharmacopoeia Commissions. 2020. Chinese pharmacopoeia. Beijing: China Medical Science and Technology Press; p. 108.
- Patel RK, Jain M. 2015. NGS QC Toolkit: a platform for quality control of next-generation sequencing data. In: Nelson K, editor. Encyclopedia of metagenomics. New York, NY: Springer.
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15(1):50. doi: 10.1186/s13007-019-0435-7.
- Xue HF, Song YY, Yang YY, Bian C, Xu L, Kang TG. 2022. The complete chloroplast genome of Pulsatilla campanella Fischer ex Krylov. (Ranunculaceae, Pulsatilla Miller). Mitochondrial DNA B Resour. 7(6):1126–1128. doi: 10.1080/23802359.2022.2087556.
- Zhai W, Duan X, Zhang R, Guo C, Li L, Xu G, Shan H, Kong H, Ren Y. 2019. Chloroplast genomic data provide new and robust insights into the phylogeny and evolution of the Ranunculaceae. Mol Phylogenet Evol. 135:12–21. doi: 10.1016/j.ympev.2019.02.024.
- Zhang W, Jiang H, Yang J, Song G, Wen D, Liu W, Jin M, Wang Q, Du Y, Sun Q, et al. 2019. A high-throughput metabolomics approach for the comprehensive differentiation of four Pulsatilla Adans herbs combined with a nontargeted bidirectional screen for rapid identification of triterpenoid saponins. Anal Bioanal Chem. 411(10):2071–2088. doi: 10.1007/s00216-019-01631-6.
- Zhang NN, Lu Y, Zhang ZQ. 2021. The complete chloroplast genome sequence of Anemone reflexa (Ranunculaceae). Mitochondrial DNA B Resour. 6(2):304–305. doi: 10.1080/23802359.2020.1860710.
- Zhang T, Xing Y, Xu L, Bao G, Zhan Z, Yang Y, Wang J, Li S, Zhang D, Kang T. 2019. Comparative analysis of the complete chloroplast genome sequences of six species of Pulsatilla Miller, Ranunculaceae. Chin Med. 14(1):53. doi: 10.1186/s13020-019-0274-5.
