Abstract
Thladiantha nudiflora Hemsl. ex F.B.Forbes & Hemsl. 1887 (Cucurbitaceae) has been widely known as a traditional medicine plant. In this study, we sequenced, assembled, and annotated the complete chloroplast genome of T. nudiflora. The chloroplast genome of T. nudiflora is 156,824 base pair (bp) in length, containing a large single-copy region of 86,566 bp and a small single-copy region of 18,070 bp, separated by a pair of inverted repeats of 26,094 bp. The chloroplast genome contains 132 genes, including 87 protein-coding, 37 transfer RNA, and eight ribosomal RNA genes. Phylogenetic analysis of the chloroplast genome revealed that species of the genus Thladiantha were clustered together in the phylogenetic trees. This study will not only shed light on T. nudiflora’s evolutionary position but also provide valuable chloroplast genomic information for future studies into the origins and diversification of the genus Thladiantha and the Cucurbitaceae family.
Introduction
Cucurbitaceae is a family of climbers or trailers native to tropical and subtropical climates, with approximately 960 accepted species (Schaefer et al. Citation2009). Thladiantha nudiflora Hemsl. ex F.B.Forbes & Hemsl. 1887 is a species belonging to the genus Thladiantha in the Cucurbitaceae family. It is native to Eastern Asia (Anh et al. Citation2020). Thladiantha nudiflora prefers sunny and fertile conditions. It is distributed mainly in South and Southeast Asia (Xu and Le Citation2017). Traditional Chinese medicine has employed Thladiantha species to soften hard masses, clear heat and detoxify, reduce swelling, and get rid of carbuncles (Nie et al. Citation1989; Wang et al. Citation2011). The first complete T. nudiflora chloroplast genome was identified and described in this article. This study offers potential genetic data for Cucurbitaceae phylogenetic and systematic molecular ecology investigations.
Materials and methods
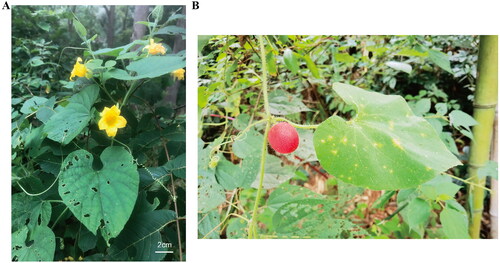
The leaves of T. nudiflora were gathered from Xuancheng, Anhui, China (GPS: 118°41′30.73″E 30°34′15.49″N). The samples were deposited at the herbarium of the College of Life Sciences and Medicine, Zhejiang Sci-Tech University, Hangzhou (Plant Genomics & Molecular Improvement of Colored Fiber Lab, http://sky.zstu.edu.cn/, Identifier: Yan-Yan Zhao, [email protected]) under voucher number ZSTUTN01. The leaves and flowers of T. nudiflora were photographed by ourselves and shown in . No permits are needed to get the sample because this species is neither endangered nor protected.
Figure 1. Species image of Thladiantha nudiflora showing the morphology of leaves and flowers (A) and fruit (B). Photograph was taken by Yan-Yan Zhao in Xuancheng, Anhui, China. Main identifying traits: Plants densely pubescent-hispid; leaves slightly stiff, blade ovate-cordate; with tendrils; female flowers solitary; flowering in spring and summer; fruiting pedicel robust, 2.5–5.5 cm; fruit red or red-brown when mature.
Using the DNA Plantzol Reagent (Invitrogen, Carlsbad, CA, USA), total genomic DNA was extracted from leaves according to the manufacturer’s recommendations. Sequencing libraries were procured as directed by the manufacturer using Illumina’s TruSeq Nano DNA Library Preparation kit (350 bp median insert). The library was sequenced on the Illumina HiSeq 2500 platform (Illumina Inc., San Diego, CA, USA). All raw readings were processed with Trimmomatic v0.39 software (Jülich, Germany) to remove adaptor sequences, short reads (length 75 bp), and low-quality bases (Q-value 20). Then, with adaptors trimmed, 21.3 million high-quality raw reads (150 bp paired-end read length) were produced. The plastid-like readings were acquired from clean reads and assembled using the GetOrganelle ver.1.7.6.1 (Jin et al. Citation2020). We further mapped our clean reads back to the assembled chloroplast genome to evaluate the depth of coverage in order to clarify the accuracy of the assembly (Supplementary Figure S1). The genome was annotated using Geneious v11.1.5 (Kearse et al. Citation2012), with the Thladiantha dubia chloroplast genome as a reference (GenBank: NC_046855). Manual corrections were made to annotation mistakes. The CPGview-RSG (http://www.1kmpg.cn/cpgview/) was used to illustrate the structural properties of the chloroplast genomes.
We received 17 available complete chloroplast genomes of Cucurbitaceae from the National Center for Biotechnology information database to corroborate the phylogenetic position of T. nudiflora. Using MAFFT v7.3 (Katoh and Standley Citation2013), the entire chloroplast genome sequence was aligned. Phylogenetic tree was constructed based on the complete chloroplast genome using maximum-likelihood (ML) and Bayesian inference (BI). ML phylogenetic tree was constructed using IQ-TREE v1.6.12 (Nguyen et al. Citation2015). The best-fitting model was determined by ModelFinder (Kalyaanamoorthy et al. Citation2017) and was GTR + F + R3 and the branch support was tested with 1000 replications. BI phylogeny was inferred using MrBayes3.2.7 (Ronquist et al. Citation2012) with the optimal model GTR + G + I. The analysis was run with the Markov chain Monte Carlo (MCMC) of 1 million generations, in which every 1000 generations were sampled and the first 25% of MCMC samples were discarded as burn-in.
Results
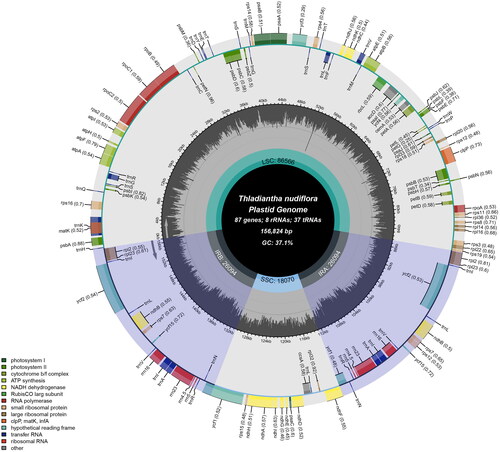
The full length of T. nudiflora chloroplast genome sequence (GenBank accession number OQ286031) was 156,824 base pairs and contained two inverted repeat (IR, 26,094 bp) sections, a large single-copy region (LSC, 86,566 bp), and a small single-copy region (SSC, 18,070 bp). The overall GC content was 37.1%, and the GC content of the IR, LSC, and SSC was 42.7%, 34.9%, and 31.2%, respectively. There were 132 genes in the genome (87 protein-coding, eight rRNA, and 37 tRNA) (). Nineteen of these genes had another copy, including eight protein-coding genes (rps7, ndhB, ycf2, rpl2, rps12, rpl23, ycf15, ycf1), seven tRNA genes (trnA-UGC, trnE-UUC, trnR-ACG, trnV-GAC, trnN-GUU, trnM-CAU, trnL-CAA), and all four rRNA genes (rrn5, rrn16, rrn4.5, rrn23). Two introns were identified in three genes (clpP, rps12, ycf3) and one intron in five protein-coding genes (rpl2, ndhB, rps16, rpoC1, ndhA) (Supplementary Figure S2).
Figure 2. The entire map of Thladiantha nudiflora’s chloroplasts. The genes in the circle are transcribed clockwise while those on the outside are transcribed counterclockwise. Genes are colored differently according to their role. In the middle circle, the GC information is displayed in a deeper shade of gray, while the AT content is displayed in a lighter shade.
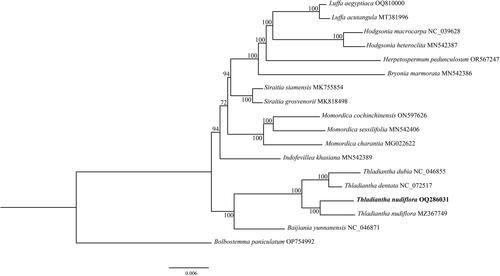
To understand the phylogenetic position of T. nudiflora in Cucurbitaceae, we downloaded complete chloroplast genomes of 17 species of the Cucurbitaceae family from the NCBI GenBank database. Most cucurbits have berry or pepo fruits (Barrera-Redondo et al. Citation2020). Since Bolbostemma paniculatum is one of the few capsule fruits in Cucurbitaceae, it was used as an outgroup to construct the phylogenetic tree. The phylogenies reconstructed by ML methods () and BI (Supplementary Figure S3) were topologically identical. Species of the genus Thladiantha were clustered together in the phylogenetic trees, suggesting this genus was a monophyletic group. Additionally, Baijiania yunnanensis exhibited as the sister clade to the species of the genus Thladiantha.
Figure 3. Phylogenetic tree of 18 species in Cucurbitaceae was constructed based on the complete chloroplast genome using maximum-likelihood (ML). Numbers at the nodes indicate bootstrap support values, and the scale bar represents nucleotide substitutions per site. Bolbostemma paniculatum was used as outgroups. The following sequences were used: Baijiania yunnanensis (NC_046871, Bellot et al. Citation2020), Bolbostemma paniculatum (OP754992), Bryonia marmorata (MN542386, Bellot et al. Citation2020), Herpetospermum pedunculosum (OR567247, Wang et al. Citation2020), Hodgsonia heteroclita (MN542387, Bellot et al. Citation2020), Hodgsonia macrocarpa (NC_039628, Zeng et al. Citation2018), Indofevillea khasiana (MN542389, Bellot et al. Citation2020), Luffa acutangula (MT381996, Yundaeng et al. Citation2020), Luffa aegyptiaca (OQ810000), Momordica charantia (MG022622), Momordica cochinchinensis (ON597626, Cai et al. Citation2023), Momordica sessilifolia (MN542406, Bellot et al. Citation2020), Siraitia grosvenorii (MK818498), Siraitia siamensis (MK755854, Shi et al. Citation2019), T. dentata (NC_072517), T. dubia (NC_046855, Bellot et al. Citation2020), T. nudiflora (MZ367749), T. nudiflora (QQ286031).
Discussion and conclusion
At present, there is little research on the phylogeny of Thladiantha, mainly because there are few chloroplast genomes published. In this study, the chloroplast genome sequence of T. nudiflora was assembled and annotated, which can be subsequently used for DNA barcoding and molecular phylogeny of Thladiantha. The complete chloroplast genome of T. dubia and Thladiantha dentata, other species of the genus Thladiantha, have been publicly available (Bellot et al. Citation2020). Phylogenetic analysis showed that species of the genus Thladiantha were clustered together in the phylogenetic trees, suggesting this genus was a monophyletic group. Additionally, Baijiania yunnanensis exhibited as the sister clade to the species of the genus Thladiantha. In conclusion, this study will not only shed light on T. nudiflora’s evolutionary position but also provide valuable chloroplast genomic information for future studies into the origins and diversification of the genus Thladiantha and the Cucurbitaceae family.
Author contributions
Yan-Yan Zhao: conception and design, analysis and interpretation of the data, drafting of the manuscript. Min-Min Chen: data analysis. Bai-Lin Duan and Qing-Zhou Xie: interpretation of data for the work and revising it critically for intellectual content. Qiang Miu: review, editing, analysis, and acquisition of data for the work and funding acquisition. All authors have agreed to be accountable for all aspects of the work.
Ethical approval
No permits are needed to get the sample because this study does not involve any endangered or protected species.
Health and safety
The authors certify that any disclosed experimental work was carried out in accordance with the necessary laboratory health and safety protocols.
Supplemental Material
Download JPEG Image (873.8 KB)Supplemental Material
Download JPEG Image (350.9 KB)Supplemental Material
Download JPEG Image (110.6 KB)Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the manuscript.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI (https://www.ncbi.nlm.nih.gov) under accession no. OQ286031. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA822536, SRR18578198, and SAMN27191640, respectively.
Additional information
Funding
References
- Anh NV, Victor D, Anh VTN, Ludmina D, Phuong DTL, Olga K. 2020. Thladiantha seed oils – new source of conjugated fatty acids: characterization of triacylglycerols and fatty acids. J Oleo Sci. 69(9):993–1000. doi:10.5650/jos.ess20075.
- Barrera-Redondo J, Lira-Saade R, Eguiarte LE. 2020. Gourds and tendrils of Cucurbitaceae: how their shape diversity, molecular and morphological novelties evolved via whole-genome duplications. Mol Plant. 13(8):1108–1110. doi:10.1016/j.molp.2020.06.012.
- Bellot S, Mitchell TC, Schaefer H. 2020. Phylogenetic informativeness analyses to clarify past diversification processes in Cucurbitaceae. Sci Rep. 10(1):488. doi:10.1038/s41598-019-57249-2.
- Cai L, Pan R, Zeng Q, Zhang X, Zeng R, Zhu Q. 2023. The complete plastome sequence of Momordica cochinchinensis (Cucurbitaceae). Mitochondrial DNA B Resour. 8(3):329–332. doi:10.1080/23802359.2023.2181649.
- Jin JJ, Yu WB, Yang JB, Song Y, dePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241. doi:10.1186/s13059-020-02154-5.
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589. doi:10.1038/nmeth.4285.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. doi:10.1093/molbev/mst010.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649. doi:10.1093/bioinformatics/bts199.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. doi:10.1093/molbev/msu300.
- Nie RL, Tanaka T, Miyakoshi M, Kasai R, Morita T, Zhou J, Tanaka O. 1989. A triterpenoid saponin from Thladiantha hookeri var. pentadactyla. Phytochemistry. 28(6):1711–1715. doi:10.1016/S0031-9422(00)97830-0.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542. doi:10.1093/sysbio/sys029.
- Schaefer H, Heibl C, Renner SS. 2009. Gourds afloat: a dated phylogeny reveals an Asian origin of the gourd family (Cucurbitaceae) and numerous oversea dispersal events. Proc Biol Sci. 276(1658):843–851. doi:10.1098/rspb.2008.1447.
- Shi H, Yang M, Mo C, Xie W, Liu C, Wu B, Ma X. 2019. Complete chloroplast genomes of two Siraitia Merrill species: comparative analysis, positive selection and novel molecular marker development. PLoS One. 14(12):e0226865. doi:10.1371/journal.pone.0226865.
- Wang C, Wang X, Tseringand T, Song Y, Zhu R. 2020. The plastid genome of Herpetospermum pedunculosum (Cucurbitaceae), an endangered traditional Tibetan medicinal herbs. Mitochondrial DNA B Resour. 5(1):495–497. doi:10.1080/23802359.2019.1703603.
- Wang L, Zhao D, Di L, Cheng D, Zhou X, Yang X, Liu Y. 2011. The anti-hyperplasia of mammary gland effect of Thladiantha dubia root ethanol extract in rats reduced by estrogen and progestogen. J Ethnopharmacol. 134(1):136–140. doi:10.1016/j.jep.2010.11.071.
- Xu Z, Le C. 2017. Identification and control of common weeds. Vol. 3. Hangzhou: Zhejiang University Press.
- Yundaeng C, Nawae W, Naktang C, Shearman JR, Sonthirod C, Sangsrakru D, Yoocha T, Jomchai N, Sheedy JR, Mekiyanon S, et al. 2020. Chloroplast genome data of Luffa acutangula and Luffa aegyptiaca and their phylogenetic relationships. Data Brief. 33:106470. doi:10.1016/j.dib.2020.106470.
- Zeng CX, Hollingsworth PM, Yang J, He ZS, Zhang ZR, Li DZ, Yang JB. 2018. Genome skimming herbarium specimens for DNA barcoding and phylogenomics. Plant Methods. 14(1):43. doi:10.1186/s13007-018-0300-0.
