Abstract
Dendrocalamus liboensis Hsueh & D. Z. Li 1985 is a unique member of the Bambusoideae subfamily found in Guizhou, China. The species has both economic importance and ornamental value. This study represents the first report of the sequencing and assembly of the complete chloroplast genome of D. liboensis. The total length of the genome was 139,483 bp, with a conventional quadripartite framework consisting of a large single-copy (LSC) region (83,001 bp in length), a small single-copy (SSC) region (12,896 bp in length), and two inverted repeats (IR) regions (both 21,793 bp in length). Overall, the D. liboensis chloroplast genome contained 128 functional genes, including 83 protein-coding genes, 37 tRNAs, and 8 rRNAs. Phylogenetic analysis showed that D. liboensis closely resembled D. sapidus, with both found on a strongly supported branch of the phylogenetic tree.
Introduction
First described in 1985, Dendrocalamus liboensis Hsueh & D. Z. Li is a member of the Dendrocalamus genus that is structurally similar to D. tsiangii but with larger rods. D. liboensis is only found in Libo County in Guizhou, China, where it is endemic and grows primarily in habitats at altitudes of about 650 m. In common with species such as Chimonobambusa lactistriata, Indocalamus hirsutissimus, and Ampelocalamus scandens, D. liboensis is increasingly threatened by deteriorating natural environmental conditions and a reduction in the size of the wild population (Zhong et al. Citation2021; Hu et al. Citation2022; Wu et al. Citation2022). Bamboo shoots serve as a wild vegetable consumed by local populations and are also used as ornamental garden plants, while bamboo culm is also valued as a building material (Du et al. Citation2009; Deng and Wang Citation2006).
Dendrocalamus is a member of the subgenus Bambusa within the Poaceae family. Li and Hsueh (Citation1988) classification divides the genus Dendrocalamus into two subgenera and five groups, whereas the Forest Code (FOC) divides it into three subgenera and seven groups. Recent systematics studies have shown that Dendrocalamus is closely related to Bambusa and Gigantochloa, which together form the BDG complex (Yang et al. Citation2008). To date, there has been no analysis of the complete chloroplast genome of D. liboensis. Thus, to contribute to the preservation of this species and explore its genetic resources by furthering research efforts, we sequenced, assembled, and analyzed the full D. liboensis chloroplast genome. The present report provides a comprehensive description of the D. liboensis chloroplast genome and its phylogenetic relationships with other Dendrocalamus species.
Materials and methods
Fresh leaves of D. liboensis () were harvested from specimens found in Libo County, Guizhou, China, in April 2022 (25°26′23.24″N, 107°55′0.26″E, altitude: 628 m). The leaves were placed in silica gel and specimens were deposited at the Natural Museum of Guizhou University (contact person: Guang-Qian Gou; email: [email protected]) under the voucher number GB264. Total DNA was extracted from the leaves using a modified CTAB method (Doyle and Doyle Citation1987), and the quality and quantity of the DNA was assessed using 1% agarose gel electrophoresis and spectrophotometry (Nanodrop, Thermo Fisher, Waltham, MA, USA), respectively.
Figure 1. Morphology and habitat map of Dendrocalamus liboensis Hsueh & D. Z. Li (These photographs were taken by Guangqian Gou in Libo, Guizhou, China). (A) Clump habit; (B) Leaf branch; (C) Internode. D. liboensis is an evergreen bamboo with 8–15 m culms, initially densely white powdery, branches usually from 6th or 7th node up, Central branch dominant and branchlets with 3–9 leaves.

The DNA samples were sent to BMK-Beijing (China) for library construction and sequencing on an Illumina HiSeq 2500 instrument (Illumina, San Diego, CA). After quality control, 4.24 Gb of clean data were obtained and assembled into circular contigs with GetOrganelle v.1.7.5 (Jin et al. Citation2020) and annotated through Plastid Genome Annotator (PGA) (Qu et al. Citation2019), using Dendrocalamus brandisii (MK679786) as the reference (Liu et al. Citation2020). Genious 9.0.2 was used for the manual correction of the annotated sequencing data (Kearse et al. Citation2012). The map of the chloroplast genome was drawn using OGDRAW v1.3.1. Mapping raw reads into the chloroplast genome sequence to generated the coverage plot by matplotlib v.3.7.0 (https://matplotlib.org/) (Supplementary Figure 1). The cis-splicing genes and trans-splicing genes were processed using CPGview (Liu et al. Citation2023). The completed D. liboensis chloroplast genome was uploaded to GenBank (accession number OR083041).
Phylogenetic relationships between D. liboensis and other Dendrocalamus species were explored by downloading 18 complete Dendrocalamus chloroplast genome sequences from the NCBI database, as well as the chloroplast genomes of Bambusa emeiensis and Bambusa multiplex as outgroups. The sequences were aligned using MAFFT (Katoh and Standley Citation2013), and the optimal model for phylogenetic tree construction was selected with IQtree (Nguyen et al. Citation2015).
Results
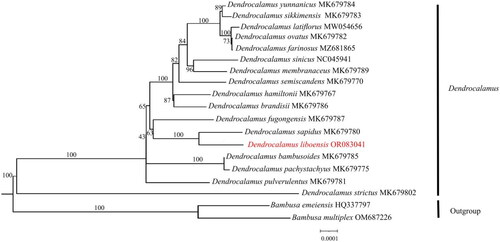
The sequencing analysis showed that the D. liboensis chloroplast genome had the typical quadrapartite structure observed in most angiosperms. At an overall length of 139,483 bp, with an average coverage depth of 257.63×(Supplementary Figure 1). It showed a typical quadripartite structure, including a large single-copy (LSC) region of 83,001 bp in length, a small single copy (SSC) region of 12,896 bp, as well as two inverse repeats (IR) segments of 21,793 bp each, with GC contents of 37.0%, 33.2%, and 44.2%, respectively (). The genome contained 128 functional genes, including 83 protein-coding genes, 37 tRNAs, and 8 rRNAs. Introns were present in 6 tRNA genes (trnK-UUU, trnI-GAU, trnA-UGC, trnG-UCC, trnV-UAC, and trnL-UAA) together with 10 protein-coding genes (atpF, ndhA, ndhB, petB, petD, rpl2, rpl16, rps16, ycf3, and rps12), with 14 genes containing one intron and two genes (ycf3, rps12) containing two introns with two copies (Supplementary Figure 2). We constructed an ML tree with 18 complete cp genomes to determine the phylogenetic position of D. liboensis. The phylogenetic analysis showed that D. liboensis was closest to D. sapidus, with both present on a highly supported branch (BS = 100%) ().
Figure 2. Map of the D. liboensis chloroplast genome(GenBank: OR083041). This map was drawn using the OGDRAW. Genes inside the circle are transcribed clockwise, and those on the outside are transcribed counter-clockwise. Genes with related functions are shown in the same color. The darker gray in the inner circle corresponds to DNA G + C content, while the lighter gray corresponds to A + T content. LSC: large single-copy; SSC: small single-copy; IR: inverted repeat.
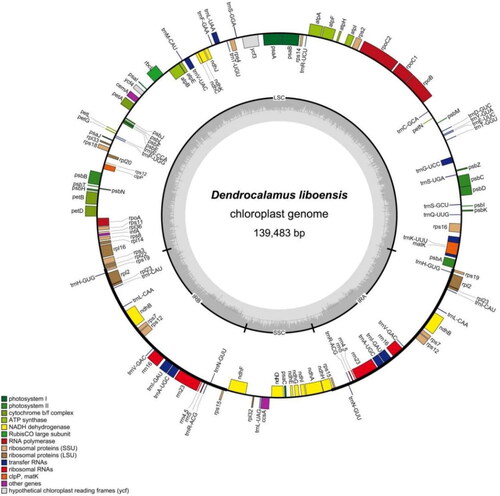
Figure 3. Maximum-likelihood(ML) phylogenetic tree of 19 species by IQtree based on complete chloroplast genomes, with Bambusa emeiensis HQ337797 (Zhang et al. Citation2011) and Bambusa multiplex OM687226 as outgroups. The phylogenetic tree was constructed using the maximum-likelihood method (ML) and bootstrap was performed 1000 times. Numbers on the nodes represent the bootstrap support values. The red fonts represents the assembled plastome sequence in this study. The following sequences were used: Dendrocalamus bambusoides MK679785 (Liu et al. Citation2020), Dendrocalamus birmanicus MK679772 (Liu et al. Citation2020), Dendrocalamus brandisii MK679786 (Liu et al. Citation2020), Dendrocalamus farinosus MZ681865, Dendrocalamus hamiltonii MK679767 (Liu et al. Citation2020), Dendrocalamus latiflorus MW054656(Liu et al. Citation2020), Dendrocalamus membranaceus MK679789 (Liu et al. Citation2020), Dendrocalamus fugongensis MK679787 (Liu et al. Citation2020), Dendrocalamus ovatus MK679782 (Liu et al. Citation2020), Dendrocalamus pachystachyus MK679775 (Liu et al. Citation2020), Dendrocalamus pulverulentus MK679781 (Liu et al. Citation2020), Dendrocalamus sapidus MK679780 (Liu et al. Citation2020), Dendrocalamus semiscandens MK679770 (Liu et al. Citation2020), Dendrocalamus sikkimensis MK679783 (Liu et al. Citation2020), Dendrocalamus sinicus NC045941 (Wang et al. Citation2019), Dendrocalamus strictus MK679802 (Liu et al. Citation2020), Dendrocalamus yunnanicus MK679784 (Liu et al. Citation2020). Undescribed citations in the legend indicate that the citations have not been published.
Discussion and conclusion
Compared with nuclear and mitochondrial genomes, chloroplast genomes are highly conserved and play an important role in phylogeny and species evolution studies (Du et al. Citation2017). With the development of high-throughput sequencing technology, chloroplast genome sequences are widely used as super barcodes in species identification and other studies (Li et al. Citation2021). In this study, the complete chloroplast genome of the economically important and ornamentally valuable species D. liboensis was sequenced for the first time and formally submitted to genome repositories. The chloroplast genome structure of D. liboensis shows a typical tetrad structure, including the LSC region, SSC region and IR region. The D. liboensis chloroplast genome was found to be similar in length and composition to those of other Dendrocalamus species (Pei et al. Citation2022). The geographical distribution of D. liboensis and D. sapidus is similar, both occurring in southern Guizhou. In addition, these two species have similar morphological characters, including bamboo stems, branches and leaves. The close relationship between the two species is consistent with the results of phylogenetic analyses using chloroplast genome construction. The result enriches the genomic data for the genus Dendrocalamus, which will contribute to phylogenetic and evolutionary studies in future.
Author contributions
Xue Xu, Mingli Wu, Guangqian Gou, Tanghang Wei, Daoping Yang and Zhaoxia Dai were involved in the conception and design. Xue Xu and Zhaoxia Dai designed the experiments. Mingli Wu, Guangqian Gou, Tanghang Wei and Daoping Yang contributed to the investigation and were involved in the collection of materials. Xue Xu and Mingli Wu were involved in the analysis and interpretation of the data. Xue Xu prepared the first draft of the manuscript. Xue Xu and Zhaoxia Dai revised the manuscript. All authors agree to be accountable for the final manuscript.
Ethical approval
The field studies were performed in line with local legislation, and appropriate permission was obtained before samples were collected from Libo County.
Supplemental Material
Download MS Word (171 KB)Acknowledgments
The authors thank other members of the laboratory for their help and suggestions for this research.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
Genome sequence data that support the findings of this study can be obtained from GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under accession no. OR083041. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA978368, SRR24793390, and SAMN35387695 respectively.
Additional information
Funding
References
- Deng LX, Wang ZJ. 2006. Study on the resources of primary bamboos and their ornamental characteristics in Guizhou province. Guizhou Forest Sci Technol. 01:48–54. CNKI:SUN:GZLY.0.2006-01-014.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15. doi:10.1016/S0021-9258(18)50745-6.
- Du XC, Ran JC, Ou ZX, Lan HB. 2009. Utilization status and protection measures of wild vegetable resources in Libo county. J Hebei Agric Sci. 13(08):15–18. doi:10.16318/j.cnki.hbnykx.2009.08.035.
- Du YP, Bi Y, Yang FP, Zhang MF, Chen XQ, Xue J, Zhang XH. 2017. Complete chloroplast genome sequences of Lilium: insights into evolutionary dynamics and phylogenetic analyses. Sci Rep. 7(1):5751. doi:10.1038/s41598-017-06210-2.
- Hu XP, Wei TL, Zhu X, Dai ZX. 2022. Population status and protection evaluation of endemic bamboo species Indocalamus hirsutissimus in Wangmo county. J Mountain Agric Biol. 41:67–70. doi:10.15958/j.cnki.sdnyswxb.2022.04.010.
- Jin JJ, Yu WB, Yang JB, Song Y, DePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241. doi:10.1186/s13059-020-02154-5.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. doi:10.1093/molbev/mst010.
- Kearse M, Moir R, Wilson A, Amy W, Stones HS, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649. doi:10.1093/bioinformatics/bts199.
- Li DZ, Hsueh CJ. 1988. A study on the genus Dendrocalamus nees from China. J Bamboo Res. 03:1–19. doi:CNKI:SUN:ZZYJ.0.1988-03-000.
- Li J, Tang J, Zeng S, Han F, Yuan J, Yu J. 2021. Comparative plastid genomics of four Pilea (Urticaceae) species: insight into interspecific plastid genome diversity in Pilea. BMC Plant Biol. 21(1):25. doi:10.1186/s12870-020-02793-7.
- Liu JX, Zhou MY, Yang GQ, Zhang YX, Ma PF, Guo C, Vorontsova MS, Li DZ. 2020. ddRAD analyses reveal a credible phylogenetic relationship of the four main genera of Bambusa-Dendrocalamus-Gigantochloa complex (Poaceae: Bambusoideae). Mol Phylogenet Evol. 146:106758. doi:10.1016/j.ympev.2020.106758.
- Liu S, Ni Y, Li J, Zhang X, Yang H, Chen H, Liu C. 2023. CPGView: a package for visualizing detailed chloroplast genome structures. Mol Ecol Resour. 23(3):694–704. doi:10.1111/1755-0998.13729.
- Nguyen LT, Schmidt HA, Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and efective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. doi:10.1093/molbev/msu300.
- Pei JL, Wang Y, Zhuo J, Gao HB, Vasupalli N, Hou D, Lin XC. 2022. Complete chloroplast genome features of Dendrocalamus farinosus and its comparison and evolutionary analysis with other Bambusoideae species. Genes (Basel). 13(9):1519. doi:10.3390/genes13091519.
- Qu X-J, Moore MJ, Li D-Z, Yi T-S. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15(1):50. doi:10.1186/s13007-019-0435-7.
- Wang Y, Yuan XL, Li YQ, Zhang JF. 2019. The complete chloroplast genome sequence of Dendrocalamus sinicus. Mitochondrial DNA B Resour. 4(2):2988–2989. doi:10.1080/23802359.2019.1664947.
- Wu ML, Wei TL, Liu YJ, Zhu X, Dai ZX. 2022. The current situation of the wild resources of the endemic species of Chimonobambusa lactistriata in Guizhou. J Mountain Agric Biol. 41:64–68. doi:10.15958/j.cnki.sdnyswxb.2022.03.009.
- Yang HQ, Yang JB, Peng ZH, Gao J, Yang YM, Peng S, Li DZ. 2008. A molecular phylogenetic and fruit evolutionary analysis of the major groups of the paleotropical woody bamboos (Gramineae: Bambusoideae) based on nuclear ITS, GBSSI gene and plastid trnL-F DNA sequences. Mol Phylogenet Evol. 48(3):809–824. doi:10.1016/j.ympev.2008.06.001.
- Zhang YJ, Ma PF, Li DZ. 2011. High-throughput sequencing of six bamboo chloroplast genomes: phylogenetic implications for temperate woody bamboos (Poaceae: Bambusoideae). PLoS One. 6(5):e20596. doi:10.1371/journal.pone.0020596.
- Zhong HM, Gou GS, Liu YJ, Zhu X, Dai ZX. 2021. Population status and protection evaluation of endemic bamboo species Ampelocalamus Scandens in Chishui river watershed. J Mountain Agric Biol. 40:71–74. doi:10.15958/j.cnki.sdnyswxb.2021.0.
